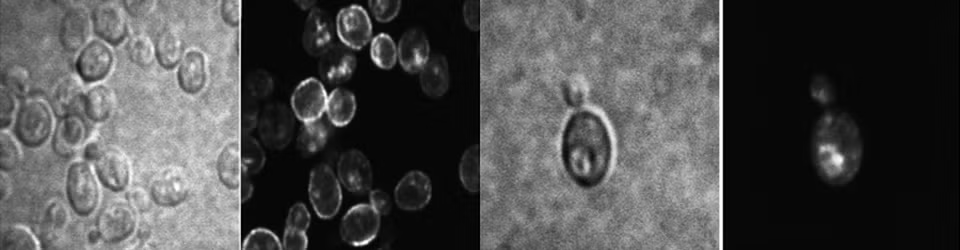
Peroxisomal support of mitochondrial respiratory efficiency promotes ER stress survival
Imadeddin Hijazi, Emily Wang, Michelle Orozco, Sarah Pelton and Amy Chang (2022) Journal of Cell Science; doi:10.1242/jcs.259254
Endoplasmic reticulum stress (ERS) occurs when cellular demand for protein folding exceeds the capacity of the organelle. Adaptation and cell survival in response to ERS requires a critical contribution by mitochondria and peroxisomes. During ERS responses, mitochondrial respiration increases to ameliorate reactive oxygen species (ROS) accumulation.We now show in yeast that peroxisome abundance also increases to promote an adaptive response. In pox1Δ cells, which are defective in peroxisomal β-oxidation of fatty acids, the respiratory response to ERS is impaired and ROS accrues. However, the respiratory response to ERS is rescued and ROS production is mitigated in pox1Δ cells overexpressing Mpc1, the mitochondrial pyruvate carrier that provides another source of acetyl CoA to fuel the tricarboxylic acid cycle and oxidative phosphorylation. Using proteomics, select mitochondrial proteins were identified that undergo upregulation upon ERS to remodel the respiratory machinery. The abundance of several peroxisome-based proteins was also increased, corroborating the role of peroxisomes in ERS adaptation. Finally, ERS stimulates assembly of respiratory complexes into higher-order supercomplexes, underlying increased electron transfer efficiency. Our results highlight peroxisomal and mitochondrial support for ERS adaptation to favor cell survival.
Retrograde signaling mediates an adaptive survival response to endoplasmic reticulum stress in Saccharomyces cerevisiae
Imadeddin Hijazi, Jeffrey Knupp and Amy Chang (2020) Journal of Cell Science; doi:10.1242/jcs.241539
One major cause of endoplasmic reticulum (ER) stress is homeostatic imbalance between biosynthetic protein folding and protein folding capacity. Cells utilize mechanisms such as the unfolded protein response (UPR) to cope with ER stress. Nevertheless, when ER stress is prolonged or severe, cell death may occur, accompanied by production of mitochondrial reactive oxygen species (ROS). Using a yeast model (Saccharomyces cerevisiae), we describe an innate, adaptive response to ER stress to increase select mitochondrial proteins, O2 consumption and cell survival. The mitochondrial response allows cells to resist additional ER stress. The ER stress induced mitochondrial response is mediated by activation of retrograde (RTG) signaling to enhance anapleurotic reactions of the tricarboxylic acid cycle. Mitochondrial response to ER stress is accompanied by inactivation of the conserved TORC1 pathway, and activation of Snf1/AMPK, the conserved energy sensor and regulator of metabolism. Our results provide new insight into the role of respiration in cell survival in the face of ER stress, and should help in developing therapeutic strategies to limit cell death in disorders linked to ER stress.
Increased mitochondrial respiration promotes survival from ER stress.
J. Knupp, P. Arvan, A. Chang (2018) Cell Death and Differentiation; https://doi.org/10.1038/s41418-018-0133-4
Protein misfolding in the endoplasmic reticulum (ER) is accompanied by adaptive cellular responses to promote cell survival. We now show that activation of mitochondrial respiration is a critical component of an adaptive ER stress response, requiring the unfolded protein response (UPR) sensor Ire1, and also calcium signaling via calcineurin. In yeast and mammalian cells lacking Ire1 or calcineurin, respiratory activation is impaired in response to ER stress; accumulation of mitochondrial reactive oxygen species (ROS) triggers cell death as abrogation of ROS by antioxidants or loss of the electron transport chain (in yeast) can rescue cells from death. Significantly, cells are rescued from ER stress-induced death by mitochondrial uncoupling by CCCP to increase O2 consumption (and increase the efficiency of electron transfer). Remarkably, genetic and pharmacologic strategies to promote mitochondrial biogenesis and increase O2 consumption also alleviate ER stress-mediated ROS and death in yeast and mammalian cells. Moreover, in a yeast genetic screen, three mitochondrial proteins Mrx9, Mrm1, and Aim19 that increase mitochondrial biogenesis were identified as high copy suppressors of ER stress-mediated cell death. Our results show that enhanced mitochondrial biogenesis, linked to improved efficiency of the electron transport chain, is a powerful strategy to block ROS accumulation and promote cell survival during ER stress in eukaryotic cells.
Sphingolipid accumulation causes mitochondrial dysregulation and cell death.
J. Knupp, F. Martinez-Montanes, F. Van Den Bergh, S. Cottier, R. Schneiter, D. Beard, A. Chang (2017) – Cell Death and Differentiation 24: 2044-2053; doi:10.1038/cdd.2017.128
Sphingolipids are structural components of cell membranes that have signaling roles to regulate many activities, including mitochondrial function and cell death. Sphingolipid metabolism is integrated with numerous metabolic networks, and dysregulated sphingolipid metabolism is associated with disease. Here, we describe a monogenic yeast model for sphingolipid accumulation. A csg2Δ mutant cannot readily metabolize and accumulates the complex sphingolipid inositol phosphorylceramide (IPC). In these cells, aberrant activation of Ras GTPase is IPC-dependent, and accompanied by increased mitochondrial reactiveoxygen species (ROS) and reduced mitochondrial mass. Survival or death of csg2Δ cells depends on nutritional status. Abnormal Ras activation in csg2Δ cells is associated with impaired Snf1/AMPK protein kinase, a key regulator of energy homeostasis. csg2Δ cells are rescued from ROS production and death by overexpression of mitochondrial catalase Cta1, abrogation of Ras hyperactivity or genetic activation of Snf1/AMPK. These results suggest that sphingolipid dysregulation compromises metabolic integrity via Ras and Snf1/AMPK pathways.
Orm proteins integrate multiple signals to maintain sphingolipid homeostasis
C. Gururaj, R. Federman, and A. Chang (2013) – J. Biol. Chem. 288: 20453-20463.
Sphingolipids are structural components of membranes, sphingolipid metabolites serve as signaling molecules. The first and rate-limiting step in sphingolipid synthesis is catalyzed by serine palmitoyltransferase (SPT). The recently discovered SPT-associated proteins, Orm1 and Orm2, are critical regulators of sphingolipids. Orm protein phosphorylation mediating feedback regulation of SPT activity occurs in response to multiple sphingolipid intermediates, including long chain base and complex sphingolipids. Both branches of the TORsignaling network, TORC1 and TORC2, participate in regulating sphingolipid synthesis via Ormp hosphorylation in response to sphingolipid >intermediates as well as nutritional conditions. Moreover,sphingolipid synthesis is regulated in response to endoplasmic reticulum (ER) stress by activation of a calcium- and calcineurin-dependent pathway via transcriptional induction of ORM2. Conversely, the calcium- and calcineurin-dependent pathway signals ER stress response upon lipid dysregulation in the absence of the Orm proteins to restore ER homeostasis.
Regulation of sphingolipid synthesis via Orm1 and Orm2 in yeast
M. Liu, C.J. Huang, S.R. Polu, R. Schneiter and A. Chang (2012) – J. Cell Sci. 125: 2428-35.
Sphingolipids are critical components of membranes and sphingolipid metabolites also serve as signaling molecules. Yeast Orm1 and Orm2 belong to a conserved family of ER membrane proteins that regulate serine palmitoyltransferase, catalyzing the first and rate-limiting step in sphingolipid synthesis. We now show that sphingolipid synthesis via Orm1 is a target of TOR signaling which regulates cell growth in response to nutritional signals. Orm1 phosphorylation is dependent on the Tap42-phosphatase complex which acts downstream of TOR protein kinase complex 1; in temperature-sensitive tap42-11 cells, impaired Orm1 phosphorylation occurs concomitantly with reduced sphingolipid synthesis. A second mechanism regulating sphingolipid synthesis is via controlling Orm2 protein level. Orm2 protein level responds to ER stress conditions, increasing when cells are treated with tunicamycin or DTT, agents that induce the unfolded protein response (UPR). The sphingolipid intermediates, long chain base and ceramide, are decreased when ORM2 is overexpressed, suggesting sphingolipid synthesis is repressed under ER stress conditions. Finally, in the absence of the Orms, the UPR is constitutively activated. Lipid dysregulation in the absence of the Orms may signal to the ER from the plasma membrane as UPR activation is dependent on a cell surface sensor and the MAPK cell wall integrity pathway. Thus, sphingolipid synthesis and the UPR are coordinately regulated. Read the Full Text
pH dependent cargo sorting from the Golgi
Chunjuan Huang and Amy Chang (2011) – J. Biol. Chem. 286: 10058-10065.
The vacuolar proton-translocating ATPase (V-ATPase) plays a major role in organelle acidification and works together with other ion transporters to maintain pH homeostasis in eukaryotic cells. We analyzed a requirement for V-ATPase activity in protein trafficking in the yeast secretory pathway. Deficiency of V-ATPase activity caused by subunit deletion or glucose deprivation results in missorting of newly synthesized plasma membrane proteins, Pma1 and Can1, directly from the Golgi to the vacuole. Vacuolar mislocalization of Pma1 is dependent on Gga adaptors although no Pma1 ubiquitination was detected. Proper cell surface targeting of Pma1 was rescued in V-ATPase deficient cells by increasing the pH of the medium, suggesting missorting is the result of aberrant cytosolic pH. In addition to mislocalization of the plasma membrane proteins, Golgi membrane proteins, Kex2 and Vrg4 are also missorted to the vacuole upon loss of V-ATPase activity. Because the missorted cargos have distinct trafficking routes, we suggest a pH dependence for multiple cargo sorting events at the Golgi. Read the Full Text
Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control.
S. Han, M.A. Lone, R. Schneiter, A. Chang (2010) – Proc. Natl. Acad. Sci. 107: 5851-6.
Yeast members of the ORMDL family of endoplasmic reticulum (ER) membrane proteins play a central role in lipid homeostasis and protein quality control. In the absence of yeast Orm1 and Orm2, accumulation of long chain base, a sphingolipid precursor, suggests dysregulation of sphingolipid synthesis. Physical interaction between Orm1 and Orm2 and serine palmitoyltransferase, responsible for the first committed step in sphingolipid synthesis, further supports a role for the Orm proteins in regulating sphingolipid synthesis. Phospholipid homeostasis is also affected in orm1Delta orm2Delta cells: the cells are inositol auxotrophs with impaired transcriptional regulation of genes encoding phospholipid biosynthesis enzymes. Strikingly, impaired growth of orm1Delta orm2Delta cells is associated with constitutive unfolded protein response, sensitivity to stress, and slow ER-to-Golgi transport. Inhibition of sphingolipid synthesis suppresses orm1Delta orm2Delta phenotypes, including ER stress, suggesting that disrupted sphingolipid homeostasis accounts for pleiotropic phenotypes. Thus, the yeast Orm proteins control membrane biogenesis by coordinating lipid homeostasis with protein quality control. Read the Full Text
A mutant plasma membrane protein is stabilized upon loss of Yvh1, a novel ribosome assembly factor
Y. Liu and A. Chang (2009) – Genetics 181: 907-915.
Pma1-10 is a mutant plasma membrane ATPase defective at the restrictive temperature in stability at the cell surface. At 37°C, Pma1-10 is ubiquitinated and internalized from the plasma membrane for degradation in the vacuole. YVH1, encoding a tyrosine phosphatase, is a mutant suppressor of pma1-10; in the absence of Yvh1, Pma1-10 remains stable at the plasma membrane, thereby permitting cells to grow. The RING finger domain of Yvh1, but not its phosphatase domain, is required for removal of mutant Pma1-10 from the plasma membrane. Yvh1 is a novel ribosome assembly factor: in yvh1Δ; cells, free 60S and 80S ribosomal subunits are decreased, free 40S subunits are increased, and half-mer polysomes are accumulated. Pma1-10 is also stabilized by deletion of 60S ribosomal proteins Rpl19 and Rpl35. We propose that changes in ribosome biogenesis caused by loss of Yvh1 or specific ribosomal proteins have effects on the plasma membrane, perhaps by producing specific translational changes. Read the Full Text
Heat shock response relieves ER stress
Y. Liu and A. Chang. (2008) – EMBO J. 27: 1049-1059.
Accumulation of misfolded protein in the endoplasmic reticulum (ER) causes stress. The unfolded protein response (UPR), a transcriptional induction pathway, is activated to relieve ER stress. Although UPR is not essential for viability, UPR-deficient cells are more sensitive to ER stress; ire1Delta cells cannot grow when challenged with tunicamycin or by overexpression of misfolded CPY(*). In these cells, multiple functions are defective, including translocation, ER-associated degradation (ERAD), and ER-to-Golgi transport. We tested whether heat shock response (HSR) can relieve ER stress. Using a constitutively active Hsf1 transcription factor to induce HSR without temperature shift, we find that HSR rescues growth of stressed ire1Delta cells, and partially relieves defects in translocation and ERAD. Cargo-specific effects of constitutively active Hsf1 on ER-to-Golgi transport are correlated with enhanced protein levels of the respective cargo receptors. In vivo, HSR is activated by ER stress, albeit to a lower level than that caused by heat. Genomic analysis of HSR targets reveals that >25% have function in common with UPR targets. We propose that HSR can relieve stress in UPR-deficient cells by affecting multiple ER activities. Read the Full Text
Cytoplasmic Hsp70 promotes ubiquitination for ER-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, Pma1
S. Han, Y. Liu, and A. Chang (2007) – J. Biol. Chem. 282: 26140-26149.
Cells have a variety of strategies for dealing with misfolded proteins. Heat shock response involves transcriptional induction of chaperones to promote and/or correct folding, and also activation of the ubiquitin/proteasome system to degrade defective proteins. In the secretory pathway, it is primarily luminal misfolded or unassembled proteins that trigger the unfolded protein response which, like heat shock, induces chaperones and components of the ER-associated degradation (ERAD) pathway. To understand cellular response to a misfolded polytopic membrane protein of the secretory pathway, we studied Pma1-D378S, a model ERAD substrate. Expression of misfolded Pma1 induces heat shock response in the absence of increased temperature. Overexpression of HSF1, the transcription factor that mediates heat shock response, increases degradation of Pma1-D378S without temperature upshift. Nevertheless, efficient Pma1-D378S degradation occurs in an hsf1 mutant that maintains basal transcription levels but cannot mediate transcriptional activation. Thus, heat shock protein induction enhances but is not necessary for ERAD. The cytoplasmic Hsp70 chaperone Ssa1 is required for ERAD of both Pma1-D378S and another transmembrane ERAD substrate, Ste6*. In the absence of Ssa chaperones, ubiquitination of both substrates is impaired, resulting in stabilization. We suggest a role for Hsp70 cytoplasmic chaperones in recognition by the ER-associated ubiquitination machinery. Read the Full Text
A role for Lte1p (a low-temperature essential protein involved in mitosis) in proprotein processing in the yeast secretory pathway
X. Zhao, A. Chang, A. Toh-e, and P. Arvan (2007) – J. Biol. Chem. 282: 1670-1678.
We previously identified 6 single gene disruptions in S. cerevisiae that allow enhanced immunoreactive insulin secretion primarily because of defective Kex2p-mediated endoproteolytic processing. Five eis mutants disrupted established vps (vacuolar protein sorting) genes, The sixth, LTE1, is a “Low-Temperature (<15°C) Essential”gene encoding a large protein with potential guanine nucleotide exchange (GEF) domains. Lte1p functions as a positive regulator of the mitotic GTPase Tem1p, and overexpression of Tem1p suppresses the low-temperature mitotic defect. By sequence analysis, Tem1p has highest similarity to Vps21p (yeast homolog of mammalian Rab5). Unlike TEM1, LTE1 is not restricted to mitosis but is expressed throughout the cell cycle. Lte1p function in interphase cells is largely unknown. Here we confirm the eis phenotype of lte1 mutant cells and demonstrate a defect in proalpha factor processing that is rescued by expression of full-length Lte1p but not a C-terminally truncated Lte1p lacking its GEF homology domain. Neither overexpression of Tem1p, nor 13 other structurally-related GTPases, can suppress the secretory proprotein processing defect. However, overexpression of Vps21p selectively restores proprotein processing in a manner dependent upon the active GTP-bound form of the GTPase. By contrast, a vps21 mutant produces a synthetic defect with lte1 in proprotein processing, as well as a synthetic growth defect. Together, the data underscore a link between the mitotic regulator, Lte1p, and protein processing and trafficking in the secretory/endosomal system. Read the Full Text
Multiple degradation pathways for misfolded mutants of the yeast plasma membrane ATPase, PMA1
Y. Liu, S. Sitaraman, and A. Chang (2006) – J. Biol. Chem. 281: 31457 – 31466.
To understand protein sorting and quality control in the secretory pathway, we have analyzed intracellular trafficking of the yeast plasma membrane ATPase, Pma1. Pma1 is ideal for such studies because it is a very abundant polytopic membrane protein, and its localization and activity at the plasma membrane are essential for cell viability and growth. We have tested whether the cytoplasmic amino and carboxy terminal domains of Pma1 carry sorting information. As the sole copy of Pma1, mutants truncated at either N- or C-termini are targeted at least partially to the plasma membrane and have catalytic activity to sustain cell viability. The mutants are also delivered to degradative pathways. Strikingly, N- and C-terminal Pma1 mutants are differentially recognized for degradation at distinct cellular locales. Cterminal mutants are recognized for destruction by ER-associated degradation. By contrast, Nterminal mutants escape detection by ERAD entirely, and undergo endocytosis for vacuolar degradation after apparently normal cell surface targeting. Both N- and C-terminal mutants are conformationally abnormal, as revealed by increased sensitivity to tryptic cleavage, but are able to assemble to form oligomers. We propose that different quality control mechanisms may assess discrete domains of Pma1 rather than a global conformational state.
Quality control of a mutant plasma membrane ATPase: ubiquitination prevents cell surface stability
Yu Liu and A. Chang (2005) – J. Cell Science 119: 360-369.
The plasma membrane ATPase, Pma1, has remarkable longevity at the cell surface. In contrast to the wild-type protein, the temperature-sensitive mutant Pma1-10 is misfolded and undergoes rapid removal from the cell surface for vacuolar degradation. At the restrictive temperature, Pma1-10 becomes ubiquitylated before or upon arrival at the plasma membrane. Internalization from the plasma membrane and vacuolar degradation of Pma1-10 is dependent on the ubiquitin-interacting motif (UIM) of the epsin Ent1, suggesting recognition of ubiquitylated substrate by the endocytic machinery. Surprisingly, ubiquitylation of Pma1-10 is reversed when its internalization is blocked in an end3 mutant. Under these conditions, Pma1-10 acquires association with detergent-insoluble, glycolipid-enriched complexes (DIGs) which has been suggested to promote stability of wild-type Pma1. Ubiquitylation does not cause DIG exclusion because a Pma1-Ub fusion protein is not significantly excluded from DIGs. We suggest that ubiquitylation of Pma1-10 represents a component of a quality control mechanism that targets the misfolded protein for removal from the plasma membrane. Rapid internalization of Pma1-10 caused by its ubiquitylation may preempt establishment of stabilizing interactions.
Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast
M. Pizzirusso and A. Chang (2004) – Mol. Biol. Cell. 15: 2401-2409.
Pma1-7 is a mutant plasma membrane ATPase which is impaired in targeting to the cell surface at 37°C and is delivered instead to the endosomal/vacuolar pathway for degradation. We have proposed that Pma1-7 is a substrate for a Golgi-based quality control mechanism. By contrast with wild-type Pma1, Pma1-7 is ubiquitinated. Ubiquitination and endosomal targeting of Pma1-7 is dependent on the Rsp5-Bul1-Bul2 ubiquitin ligase protein complex but not the transmembrane ubiquitin ligase Tul1. Analysis of Pma1-7 ubiquitination in mutants blocked in protein transport at various steps of the secretory pathway suggests that ubiquitination occurs after ER exit but before endosomal entry. In the absence of ubiquitination in rsp5-1 cells, Pma1-7 is delivered to the cell surface and remains stable. Nevertheless, Pma1-7 remains impaired in association with detergent-insoluble glycolipid-enriched complexes (DIGs) in rsp5-1 cells, suggesting that ubiquitination is not the cause of Pma1-7 exclusion from rafts. In vps1 cells in which protein transport into the endosomal pathway is blocked, Pma1-7 is routed to the cell surface. On arrival at the plasma membrane in vps1 cells, Pma1-7 remains stable and its ubiquitination disappears, suggesting deubiquitination activity at the cell surface. We suggest that Pma1-7 sorting and fate are regulated by ubiquitination.
Substrate recognition in ER-associated degradation mediated by Eps1, a member of the protein disulfide isomerase family
Q. Wang and A. Chang (2003) – EMBO J. 22: 3792-3802.
Pma1-D378N is a misfolded plasma membrane protein in yeast that is prevented from delivery to the cell surface and targeted instead for ER-associated degradation (ERAD). Degradation of Pma1-D378N is dependent on the ubiquitin ligase Doa10 and the ubiquitin chaperone Cdc48. Recognition of Pma1-D378N by the ERAD pathway is dependent on Eps1, a transmembrane member of the protein disulfide isomerase (PDI) oxidoreductase family. Eps1 has two thioredoxin-like domains containing a CPHC and a CDKC active site. Although Eps1 interaction with wild-type Pma1 was not detected, Eps1 co-immunoprecipitates with Pma1-D378N. Eps1 interaction with Pma1-D378N requires the CPHC motif, although both thioredoxin-like domains appear to cooperate in substrate recognition. In the absence of the native transmembrane domain and cytoplasmic tail of Eps1, degradation of Pma1-D378N is slowed, suggesting that Eps1 facilitates presentation of substrate to membrane-bound components of the degradation machinery. Genetic interactions with other mutants of the ERAD machinery and induction of the unfolded protein response in eps1 cells support a general role for Eps1 as a recognition component of the ERAD pathway.
Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1
Q. Wang and A. Chang (2002) – Proc. Natl. Acad. Sci. 99: 12853-12858.
The plasma membrane H+-ATPase, Pma1, is an essential and long-lived integral membrane protein. Previous work has demonstrated that the Pma1-D378N mutant is a substrate for endoplasmic reticulum (ER)-associated degradation and causes a dominant negative effect on cell growth by preventing ER export of wild-type Pma1. We now show that Pma1-D378N is ubiquitylated, and it heterooligomerizes with wild-type Pma1, resulting in ubiquitylation and ER-associated degradation of wild-type Pma1. In temperature-sensitive lcb1-100 cells, defective in sphingoid base synthesis, Pma1 fails to oligomerize. At 30°C, lcb1-100 is a suppressor of pma1-D378N because wild-type Pma1 fails to heterooligomerize with Pma1-D378N; wild-type Pma1 moves to the cell surface, indicating that oligomerization is not required for delivery to the plasma membrane. Even in the absence of Pma1-D378N, wild-type Pma1 is ubiquitylated and it undergoes internalization from the cell surface and vacuolar degradation at 30°C in lcb1-100 cells. At 37°C in lcb1-100 cells, a more severe defect occurs in sphingoid base synthesis, and targeting of newly synthesized Pma1 to the plasma membrane is impaired. These data indicate requirements for sphingolipids at three discrete stages: Pma1 oligomerization at the ER, targeting to the plasma membrane, and stability at the cell surface.
An ER membrane protein, Sop4, facilitates ER export of the yeast plasma membrane [H+]ATPase
W.-J. Luo, Xiao-hua Gong, and A. Chang (2002) – Traffic 3: 730-739.
We have analyzed the mechanism by which Sop4, a novel ER membrane protein, regulates quality control and intracellular transport of Pma1-7, a mutant plasma membrane ATPase. At the restrictive temperature, newly synthesized Pma1-7 is targeted for vacuolar degradation instead of being correctly delivered to the cell surface. Loss of Sop4 at least partially corrects vacuolar mislocalization, allowing Pma1-7 routing to the plasma membrane. Ste2-3 is a mutant pheromone receptor which, like Pma1-7, is defective in targeting to the cell surface, resulting in a mating defect. sop4delta suppresses the mating defect of ste2-3 cells as well as the growth defect of pma1-7. Visualization of newly synthesized Pma1-7 in sop4delta cells by indirect immunofluorescence reveals delayed export from the ER. Similarly, ER export of wild-type Pma1 is delayed in the absence of Sop4 although intracellular transport of Gas1 and CPY is unaffected. These observations suggest a model in which a selective increase in ER residence time for Pma1-7 may allow it to achieve a more favorable conformation for subsequent delivery to the plasma membrane. In support of this model, newly synthesized Pma1-7 is also routed to the plasma membrane upon release from a general block of ER-to-Golgi transport in sec13-1 cells.
Secretory pathway quality control in the Golgi, plasmalemmal, and endosomal systems
P. Arvan, Xiang Zhao, Jose Ramos-Castaneda, and A. Chang (2002) – Traffic 3: 771-780.
Exportable proteins that have significant defects in nascent polypeptide folding or subunit assembly are frequently retained in the endoplasmic reticulum and subject to endoplasmic reticulum-associated degradation by the ubiquitin-proteasome system. In addition to this, however, there is growing evidence for post-endoplasmic reticulum quality control mechanisms in which mutant or non-native exportable proteins may undergo anterograde transport to the Golgi complex and post-Golgi compartments before intracellular disposal. In some instances, these proteins may undergo retrograde transport back to the endoplasmic reticulum with re-targeting to the endoplasmic reticulum-associated degradation pathway; in other typical cases, they are targeted into the endosomal system for degradation by vacuolar/lysosomal proteases. Such quality control targeting is likely to involve recognition of features more commonly expressed in mutant proteins, but may also be expressed by wild-type proteins, especially in cells with perturbation of local environments that are essential for normal protein trafficking and stability in the secretory pathway and at the cell surface.
Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast
M. Bagnat, A. Chang, and K. Simons (2001) – Mol. Biol. Cell 12: 4129-4138.
Correct sorting of proteins is essential to generate and maintain the identity and function of the different cellular compartments. In this study we demonstrate the role of lipid rafts in biosynthetic delivery of Pma1p, the major plasma membrane proton ATPase, to the cell surface. Disruption of rafts led to mistargeting of Pma1p to the vacuole. Conversely, Pma1-7, an ATPase mutant that is mistargeted to the vacuole, was shown to exhibit impaired raft association. One of the previously identified suppressors, multicopy AST1, not only restored surface delivery but also raft association of Pma1-7. Ast1p, which is a peripheral membrane protein, was found to directly interact with Pma1p inducing its clustering into a SDS/Triton X100-resistant oligomer. We suggest that clustering facilitates partition of Pma1p into rafts and transport to the cell surface.
A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the cell surface
X.-H. Gong and A. Chang (2001) – Proc. Natl. Acad. Sci 98: 9104-9109.
Pma1 is a plasma membrane H+-ATPase whose activity at the cell surface is essential for cell viability. In this paper we describe a temperature-sensitive pma1 allele, pma1-10 (with two point mutations in the first cytoplasmic loop of Pma1), in which the newly synthesized mutant protein fails to remain stable at the cell surface at 37°C. Instead, Pma1-10 appears to undergo internalization for vacuolar degradation in a manner dependent on End4, Vps27, Doa4, and Pep4. By contrast with wild-type Pma1, mutant Pma1-10 is hypophosphorylated and fails to associate with a Triton-insoluble fraction at 37°C, suggesting failure to enter lipid rafts. Kinetic analysis reveals that, at the permissive temperature, newly synthesized Pma1-10 acquires Triton-insolubility before becoming stabilized. We suggest that phosphorylation and lipid raft association may play important roles in maintaining protein stability at the plasma membrane.
Intracellular retention of newly-synthesized insulin in yeast is caused by endoproteolytic processing in the Golgi complex
B.-Y. Zhang, A. Chang, T. Kjeltsen, and P. Arvan (2001) – J. Cell Biol 153:1187-1197.
An insulin-containing fusion protein (ICFP, encoding the yeast prepro- factor leader peptide fused via a lysine-arginine cleavage site to a single chain insulin) has been expressed in Saccharomyces cerevisiae where it is inefficiently secreted. Single gene disruptions have been identified that cause enhanced immunoreactive insulin secretion (eis). Five out of six eis mutants prove to be vacuolar protein sorting (vps)8, vps35, vps13, vps4, and vps36, which affect Golgiendosome trafficking. Indeed, in wild-type yeast insulin is ultimately delivered to the vacuole, whereas vps mutants secrete primarily unprocessed ICFP. Disruption of KEX2, which blocks intracellular processing to insulin, quantitatively reroutes ICFP to the cell surface, whereas loss of the Vps10p sorting receptor is without effect. Secretion of unprocessed ICFP is not based on a dominant secretion signal in the -leader peptide. Although insulin sorting mediated by Kex2p is saturable, Kex2p functions not as a sorting receptor but as a protease: replacement of Kex2p by truncated secretory Kex2p (which travels from Golgi to cell surface) still causes endoproteolytic processing and intracellular insulin retention. Endoproteolysis promotes a change in insulin’s biophysical properties. B5His residues normally participate in multimeric insulin packing; a point mutation at this position permits ICFP processing but causes the majority of processed insulin to be secreted. The data argue that multimeric assembly consequent to endoproteolytic maturation regulates insulin sorting in the secretory pathway.
An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast
W.-J. Luo and A. Chang (2000) – Mol. Biol. Cell 11:579-592.
The plasma membrane ATPase, encoded by PMA1, is delivered to the cell surface via the secretory pathway. Previously, we characterized a temperature-sensitive pma1 mutant in which newly synthesized Pma1-7 is not delivered to the plasma membrane but is mislocalized instead to the vacuole at 37°C. Several vps mutants, which are defective in vacuolar protein sorting, suppress targeting-defective pma1 by allowing mutant Pma1 to move once again to the plasma membrane. In this study, we have analyzed trafficking in the endosomal system by monitoring the movement of Pma1-7 in vps36, vps1, and vps8 mutants. Upon induction of expression, mutant Pma1 accumulates in the prevacuolar compartment in vps36 cells. After chase, a fraction of newly synthesized Pma1-7 is delivered to the plasma membrane. In both vps1 and vps8 cells, newly synthesized mutant Pma1 appears in small punctate structures before arrival at the cell surface. Nevertheless, biosynthetic membrane traffic appears to follow different routes in vps8 and vps1: the vacuolar protein-sorting receptor Vps10p is stable in vps8 but not in vps1. Furthermore, a defect in endocytic delivery to the vacuole was revealed in vps8 (and vps36) but not vps1 by endocytosis of the bulk membrane marker FM 4-64. Moreover, in vps8 cells, there is defective down-regulation from the cell surface of the mating receptor Ste3, consistent with persistent receptor recycling from an endosomal compartment to the plasma membrane. These data support a model in which mutant Pma1 is diverted from the Golgi to the surface in vps1 cells. We hypothesize that in vps8 and vps36, in contrast to vps1, mutant Pma1 moves to the surface via endosomal intermediates, implicating an endosome-to-surface traffic pathway.
Eps1, a novel PDI-related protein involved in ER quality control in yeast
Q. Wang and A. Chang (1999) – EMBO J 18: 5972-5982.
PMA1 is an essential gene encoding the yeast plasma membrane [H(+)]ATPase. A pma1-D378N mutant has a dominant-negative effect on cell growth because both newly synthesized mutant and wild-type Pma1 molecules are retained and degraded in the endoplasmic reticulum (ER). Like other substrates for ER-associated degradation, Pma1-D378N is stabilized in mutants defective in components of the ubiquitination machinery. A genetic selection was performed for eps (ER-retained pma1 suppressing) mutants in which the growth defect caused by the D378N allele is suppressed. In an eps1 mutant, both mutant and wild-type Pma1 molecules are allowed to travel to the plasma membrane; however, normal retention of resident ER proteins Shr3 and Kar2 is not perturbed. Eps1 is a novel membrane protein belonging to the protein disulfide isomerase (PDI) family, and Eps1 co-localizes with Pma1-D378N in the ER. In the absence of Pma1-D378N, ER export of wild-type Pma1 is not affected by eps1 deletion, but export of the plasma membrane protein Gas1 is delayed. Because Eps1 is required for retention and degradation of Pma1-D378N, we propose a model in which Eps1 acts as a novel membrane-bound chaperone in ER quality control.
Novel genes regulating endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant
W.-J. Luo and A. Chang (1997) – J. Cell Biol 138: 731-746.
A novel genetic selection was used to identify genes regulating traffic in the yeast endosomal system. We took advantage of a temperature-sensitive mutant in PMA1, encoding the plasma membrane ATPase, in which newly synthesized Pma1 is mislocalized to the vacuole via the endosome. Diversion of mutant Pma1 from vacuolar delivery and rerouting to the plasma membrane is a major mechanism of suppression of pma1(ts). 16 independent suppressor of pma1 (sop) mutants were isolated. Identification of the corresponding genes reveals eight that are identical with VPS genes required for delivery of newly synthesized vacuolar proteins. A second group of SOP genes participates in vacuolar delivery of mutant Pma1 but is not essential for delivery of the vacuolar protease carboxypeptidase Y. Because the biosynthetic pathway to the vacuole intersects with the endocytic pathway, internalization of a bulk membrane endocytic marker FM 4-64 was assayed in the sop mutants. By this means, defective endosome-to-vacuole trafficking was revealed in a subset of sop mutants. Another subset of sop mutants displays perturbed trafficking between endosome and Golgi: impaired pro-alpha factor processing in these strains was found to be due to defective recycling of the trans-Golgi protease Kex2. One of these strains defective in Kex2 trafficking carries a mutation in SOP2, encoding a homologue of mammalian synaptojanin (implicated in synaptic vesicle endocytosis and recycling). Thus, cell surface delivery of mutant Pma1 can occur as a consequence of disturbances at several different sites in the endosomal system.