
Primary Research
Wilinski D, Dus M. N6-adenosine methylation controls the translation of insulin mRNA. Nature Structural & Molecular Biology. 2023 Jul 24
Control of insulin mRNA translation is crucial for energy homeostasis, but the mechanisms remain largely unknown. We discovered that insulin mRNAs across invertebrates, vertebrates and mammals feature the modified base N6-methyladenosine (m6A). In flies, this RNA modification enhances insulin mRNA translation by promoting the association of the transcript with polysomes. Depleting m6A in Drosophila melanogaster insulin 2 mRNA (dilp2) directly through specific 3′ untranslated region (UTR) mutations, or indirectly by mutating the m6A writer Mettl3, decreases dilp2 protein production, leading to aberrant energy homeostasis and diabetic-like phenotypes. Together, our findings reveal adenosine mRNA methylation as a key regulator of insulin protein synthesis with notable implications for energy balance and metabolic disease.

Sung H, Vaziri A, Wilinski D, Woerner RKR, Freddolino PL, Dus M. Nutrigenomic Regulation of Sensory Plasticity. eLife. March 2023

Diet profoundly influences brain physiology, but how metabolic information is transmuted into neural activity and behavior changes remains elusive. Here, we show that the metabolic enzyme O-GlcNAc Transferase (OGT) moonlights on the chromatin of the D. melanogaster gustatory neurons to instruct changes in chromatin accessibility and transcription that underlie sensory adaptations to a high-sugar diet. OGT works synergistically with the Mitogen Activated Kinase/Extracellular signal Regulated Kinase (MAPK/ERK) rolled and its effector stripe (also known as EGR2 or Krox20) to integrate activity information. OGT also cooperates with the epigenetic silencer Polycomb Repressive Complex 2.1 (PRC2.1) to decrease chromatin accessibility and repress transcription in the high-sugar diet. This integration of nutritional and activity information changes the taste neurons’ responses to sugar and the flies’ ability to sense sweetness. Our findings reveal how nutrigenomic signaling generates neural activity and behavior in response to dietary changes in the sensory neurons.
Pardo-Garcia TR, Gu K, Woerner RKR, Dus M. Food memory circuits regulate eating and energy balance. Current Biology. 2023 Jan 23; 33, 215–227

In mammals, learning circuits play an essential role in energy balance by creating associations between sensory cues and the rewarding qualities of food. This process is altered by diet-induced obesity, but the causes and mechanisms are poorly understood. Here, we exploited the relative simplicity and wealth of knowledge about the D. melanogaster reinforcement learning network, the mushroom body, in order to study the relationship between the dietary environment, dopamine-induced plasticity, and food associations. We show flies that are fed a high-sugar diet cannot make associations between sensory cues and the rewarding properties of sugar. This deficit was caused by diet exposure, not fat accumulation, and specifically by lower dopamine-induced plasticity onto mushroom body output neurons (MBONs) during learning. Importantly, food memories dynamically tune the output of MBONs during eating, which instead remains fixed in sugar-diet animals. Interestingly, manipulating the activity of MBONs influenced eating and fat mass, depending on the diet. Altogether, this work advances our fundamental understanding of the mechanisms, causes, and consequences of the dietary environment on reinforcement learning and ingestive behavior.
Wilinski D, Dus M. N6-Adenosine methylation regulates the translation of insulin mRNA. biorxiv. 2022 Sep 20
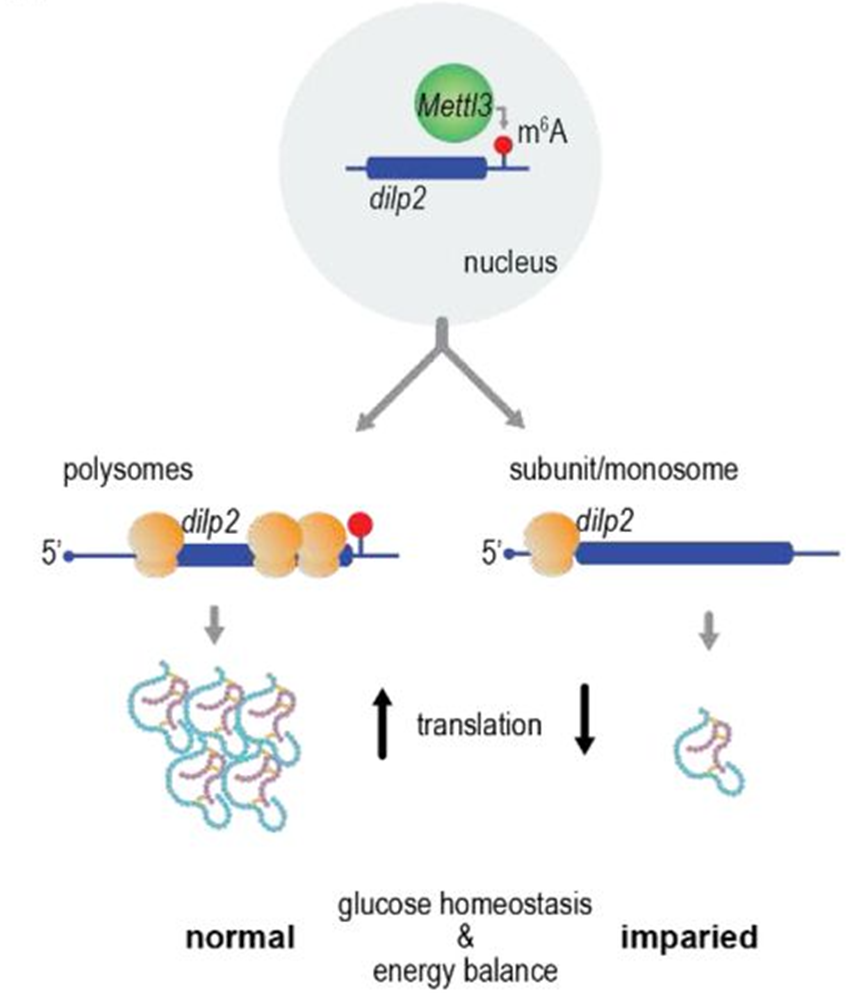
Relatively little is known about the first step of insulin synthesis: the translation of the mRNA. Here we show that the translation of D. melanogaster insulin 2 mRNA (dilp2) is controlled by methylation of N6-adenosine (m6A) in the 3’ UTR. Mutations in the m6A writer Mettl3 and methylated-residues in the dilp2 3’UTR decreased the levels of dilp2 mRNA associated with the polysomes, and the total amount of dilp2 protein produced. This resulted in aberrant energy homeostasis and diabetic-like phenotypes, consistent with the specific function of dilp2 in adult metabolism. Conserved m6A signatures were also identified in the 3’ UTRs of vertebrate insulin mRNAs. These data identify m6A as a key regulator of insulin protein synthesis and energy homeostasis in metazoans and demonstrate an essential role for m6A in translation, with important implications for diabetes and metabolic disease.
Sung H***, Vesela I***, Dricks H, Ferrario CR, Mistretta CM, Bradley RM*, Dus M*. High-sucrose diet exposure is associated with selective and reversible alterations in the rat peripheral taste system. Current Biology. August 2022.
Elevated sugar consumption is associated with an increased risk for metabolic diseases. Whereas evidence from humans, rodents, and insects suggests that dietary sucrose modifies sweet taste sensation, understanding of peripheral nerve or taste bud alterations is sparse. To address this, male rats were given access to 30% liquid sucrose for 4 weeks (sucrose rats). Neurophysiological responses of the chorda tympani (CT) nerve to lingual stimulation with sugars, other taste qualities, touch, and cold were then compared with controls (access to water only). Morphological and immunohistochemical analyses of fungiform papillae and taste buds were also conducted. Sucrose rats had substantially decreased CT responses to 0.15–2.0 M sucrose compared with controls. In contrast, effects were not observed for glucose, fructose, maltose, Na saccharin, NaCl, organic acid, or umami, touch, or cold stimuli. Whereas taste bud number, size, and innervation volume were unaffected, the number of PLCβ2+ taste bud cells in the fungiform papilla was reduced in sucrose rats. Notably, the replacement of sucrose with water resulted in a complete recovery of all phenotypes over 4 weeks. The work reveals the selective and modality-specific effects of sucrose consumption on peripheral taste nerve responses and taste bud cells, with implications for nutrition and metabolic disease risk.
Vaziri A, Morteza K, Genaw B, Freddolino PL, Dus, M*. Persistent Epigenetic Reprogramming of Sweet Taste by Diet. Science Advances. 2020 Nov 11;6(46):eabc8492. PMID: 33177090

Diets rich in sugar, salt, and fat alter taste perception and food preference, contributing to obesity and metabolic disorders, but the molecular mechanisms through which this occurs are unknown. Here, we show that in response to a high sugar diet, the epigenetic regulator Polycomb Repressive Complex 2.1 (PRC2.1) persistently reprograms the sensory neurons of Drosophila melanogaster flies to reduce sweet sensation and promote obesity. In animals fed high sugar, the binding of PRC2.1 to the chromatin of the sweet gustatory neurons is redistributed to repress a developmental transcriptional network that modulates the responsiveness of these cells to sweet stimuli, reducing sweet sensation. Half of these transcriptional changes persist despite returning the animals to a control diet, causing a permanent decrease in sweet taste. Our results uncover a new epigenetic mechanism that, in response to the dietary environment, regulates neural plasticity and feeding behavior to promote obesity.
Press release Epigenetics Podcast Episode
Wilinski D., Winzeler J., Duren W., Persons J. L., Holme K. J., Mosquera J., Khabiri M., Kinchen J. M., Freddolino P. L., Karnovsky A., Dus M*. Rapid metabolic shifts occur during the transition between hunger and satiety in Drosophila melanogaster. Nat Commun. 10, 4052 (2019).
Metabolites are active controllers of cellular physiology, but their role in complex behaviors is less clear. Here we report metabolic changes that occur during the transition between hunger and satiety in Drosophila melanogaster. To analyze these data in the context of fruit fly metabolic networks, we developed Flyscape, an open-access tool. We show that in response to eating, metabolic profiles change in quick, but distinct ways in the heads and bodies. Consumption of a high sugar diet dulls the metabolic and behavioral differences between the fasted and fed state, and reshapes the way nutrients are utilized upon eating. Specifically, we found that high dietary sugar increases TCA cycle activity, alters neurochemicals, and depletes 1-carbon metabolism and brain health metabolites N-acetyl-aspartate and kynurenine. Together, our work identifies the metabolic transitions that occur during hunger and satiation, and provides a platform to study the role of metabolites and diet in complex behavior.
May CE., Rosander J., Dennis E, Gottfried J., Dus, M*. Dietary sugar inhibits satiation by decreasing the central processing of sweet taste. eLife 2020;9:e54530. PMID: 32539934

From humans to vinegar flies, exposure to diets rich in sugar and fat lowers taste sensation, changes food choices, and promotes feeding. However, how these peripheral alterations influence eating is unknown. Here we used the genetically tractable organism D. melanogaster to define the neural mechanisms through which this occurs. We characterized a population of protocerebral anterior medial dopaminergic neurons (PAM DANs) that innervates the β’2 compartment of the mushroom body and responds to sweet taste. In animals fed a high sugar diet, the response of PAM-β’2 to sweet stimuli was reduced and delayed, and sensitive to the strength of the signal transmission out of the sensory neurons. We found that PAM-β’2 DANs activity controls feeding rate and satiation: closed-loop optogenetic activation of β’2 DANs restored normal eating in animals fed high sucrose. These data argue that diet-dependent alterations in taste weaken satiation by impairing the central processing of sensory signals.
Press release eLife Digest, When too Much Sugar is not Enough Naked Scientist Podcast
May C.E., Vaziri A, Lin Y.Q., Grushko O, Khabiri M, Wang Q-P, Holme K.J., Pletcher S.D., Freddolino P.L., Neely G.G., Dus M*. High Dietary Sugar Reshapes Sweet Taste to Promote Feeding Behavior in Drosophila melanogaster. Cell Rep. 2019 May 7; 1675-85.
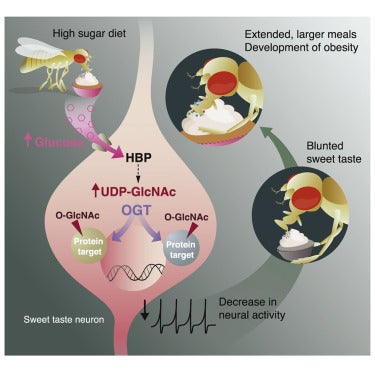
Recent studies find that sugar tastes less intense to humans with obesity, but whether this sensory change is a cause or a consequence of obesity is unclear. To tackle this question, we study the effects of a high sugar diet on sweet taste sensation and feeding behavior in Drosophila melanogaster. On this diet, fruit flies have lower taste responses to sweet stimuli, overconsume food, and develop obesity. Excess dietary sugar, but not obesity or dietary sweetness alone, caused taste deficits and overeating via the cell-autonomous action of the sugar sensor O-linked N-Acetylglucosamine (O-GlcNAc) transferase (OGT) in the sweet-sensing neurons. Correcting taste deficits by manipulating the excitability of the sweet gustatory neurons or the levels of OGT protected animals from diet-induced obesity. Our work demonstrates that the reshaping of sweet taste sensation by excess dietary sugar drives obesity and highlights the role of glucose metabolism in neural activity and behavior.
Park J-Y, Dus M, Kim S, Abu F, Kanai M.I., Rudy B, Suh G.S.B. Drosophila SLC5A11 Mediates Hunger by Regulating K+ Channel Activity. Curr Biol. 2016 Aug 8; 26 (18): 1965-74.
Hunger is a powerful drive that stimulates food intake. Yet, the mechanism that determines how the energy deficits that result in hunger are represented in the brain and promote feeding is not well understood. We previously described SLC5A11—a sodium/solute co-transporter-like—(or cupcake) in Drosophila melanogaster, which is required for the fly to select a nutritive sugar over a sweeter nonnutritive sugar after periods of food deprivation. SLC5A11 acts on approximately 12 pairs of ellipsoid body (EB) R4 neurons to trigger the selection of nutritive sugars, but the underlying mechanism is not understood. Here, we report that the excitability of SLC5A11-expressing EB R4 neurons increases dramatically during starvation and that this increase is abolished in the SLC5A11 mutation. Artificial activation of SLC5A11-expresssing neurons is sufficient to promote feeding and hunger-driven behaviors; silencing these neurons has the opposite effect. Notably, SLC5A11 transcript levels in the brain increase significantly when flies are starved and decrease shortly after starved flies are refed. Furthermore, expression of SLC5A11 is sufficient for promoting hunger-driven behaviors and enhancing the excitability of SLC5A11-expressing neurons. SLC5A11 inhibits the function of the Drosophila KCNQ potassium channel in a heterologous expression system. Accordingly, a knockdown of dKCNQ expression in SLC5A11-expressing neurons produces hunger-driven behaviors even in fed flies, mimicking the overexpression of SLC5A11. We propose that starvation increases SLC5A11 expression, which enhances the excitability of SLC5A11-expressing neurons by suppressing dKCNQ channels, thereby conferring the hunger state.
Wong JM, Malec PA, Mabrouk OS, Ro J, Dus M, Kennedy RT. Benzoyl chloride derivatization with liquid chromatography-mass spectrometry for targeted metabolomics of neurochemicals in biological samples. Journal of Chromatography A. 2016 May 13; 1446:78-90.
Widely targeted metabolomic assays are useful because they provide quantitative data on large groups of related compounds. We report a high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) method that utilizes benzoyl chloride labeling for 70 neurologically relevant compounds, including catecholamines, indoleamines, amino acids, polyamines, trace amines, antioxidants, energy compounds, and their metabolites. The method includes neurotransmitters and metabolites found in both vertebrates and insects. This method was applied to analyze microdialysate from rats, human cerebrospinal fluid, human serum, fly tissue homogenate, and fly hemolymph, demonstrating its broad versatility for multiple physiological contexts and model systems. Limits of detection for most assayed compounds were below 10nM, relative standard deviations were below 10%, and carryover was less than 5% for 70 compounds separated in 20min, with a total analysis time of 33min. This broadly applicable method provides robust monitoring of multiple analytes, utilizes small sample sizes, and can be applied to diverse matrices. The assay will be of value for evaluating normal physiological changes in metabolism in neurochemical systems. The results demonstrate the utility of benzoyl chloride labeling with HPLC-MS/MS for widely targeted metabolomics assays.
Dus M, Jason Sih-Yu Lai, Keith M. Gunapala, Soohong Min, Timothy D. Tayler, Anne C. Hergarden, Eliot Geraud, Christina M. Joseph, Greg S.B. Suh. Nutrient Sensor in the Brain Directs the Action of the Brain-Gut Axis in Drosophila. Neuron. 2015 June 11.

Animals can detect and consume nutritive sugars without the influence of taste. However, the identity of the taste-independent nutrient sensor and the mechanism by which animals respond to the nutritional value of sugar are unclear. Here, we report that six neurosecretory cells in the Drosophila brain that produce Diuretic hormone 44 (Dh44), a homolog of the mammalian corticotropin-releasing hormone (CRH), were specifically activated by nutritive sugars. Flies in which the activity of these neurons or the expression of Dh44 was disrupted failed to select nutritive sugars. Manipulation of the function of Dh44 receptors had a similar effect. Notably, artificial activation of Dh44 receptor-1 neurons resulted in proboscis extensions and frequent episodes of excretion. Conversely, reduced Dh44 activity led to decreased excretion. Together, these actions facilitate ingestion and digestion of nutritive foods. We propose that the Dh44 system directs the detection and consumption of nutritive sugars through a positive feedback loop.
Dus M, Ai M, Suh GS. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute co-transporter in Drosophila. Nature Neuroscience. 2013 Mar 31. doi: 10.1038/nn.3372.
Animals can determine the nutritional value of sugar without the influence of taste. We examined a Drosophila mutant that is insensitive to the nutritional value of sugars, responding only to the concentration (that is, sweetness). The affected gene encodes a sodium/solute co-transporter–like protein, designated SLC5A11 (or cupcake), which is structurally similar to mammalian sodium/glucose co-transporters that transport sugar across the intestinal and renal lumen. However, SLC5A11 was prominently expressed in 10 13 pairs of R4 neurons of the ellipsoid body in the brain and functioned in these neurons for
selecting appropriate foods.
Dus M, Min S, Keene AC, Lee GY, Suh GS. Taste-independent detection of the caloric content of sugars in Drosophila. Proc Natl Acad Sci U S A. 2011 Jul 12; 108 (28): 11644–11649.

Feeding behavior is influenced primarily by two factors: nutritional needs and food palatability. However, the role of food deprivation and metabolic needs in the selection of appropriate food is poorly understood. Here, we show that the fruit fly, Drosophila melanogaster, selects calorie-rich foods following prolonged food deprivation in the absence of taste-receptor signaling. Flies mutant for the sugar receptors Gr5a and Gr64a cannot detect the taste of sugar, but still consumed sugar over plain agar after 15 h of starvation. Similarly, pox-neuro mutants that are insensitive to the taste of sugar preferentially consumed sugar over plain agar upon starvation. Moreover, when given a choice between metabolizable sugar (sucrose or D-glucose) and nonmetabolizable (zero-calorie) sugar (sucralose or L-glucose), starved Gr5a; Gr64a double mutants preferred metabolizable sugars. These findings suggest the existence of a taste-independent metabolic sensor that functions in food selection. The preference for calorie-rich food correlates with a decrease in the two main hemolymph sugars, trehalose and glucose, and in glycogen stores, indicating that this sensor is triggered when the internal energy sources are depleted. Thus, the need to replenish depleted energy stores during periods of starvation may be met through the activity of a taste-independent metabolic sensing
pathway.
Keene AC, Duboué ER, McDonald DM, Dus M, Suh GSB, Waddell S, Blau J. Clock and Cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010 Jul 13; 20(13):1209-15.
Neural systems controlling the vital functions of sleep and feeding in mammals are tightly interconnected: sleep deprivation promotes feeding, whereas starvation suppresses
sleep. Here we show that starvation in Drosophila potently suppresses sleep, suggesting that these two homeostatically regulated behaviors are also integrated in flies. The sleep-suppressing effect of starvation is independent of the mushroom bodies, a previously identified sleep locus in the fly brain, and therefore is regulated by distinct neural circuitry. The circadian clock genes Clock (Clk) and cycle (cyc) are critical for proper sleep suppression during starvation. However, the sleep suppression is independent of light cues and of circadian rhythms as shown by the fact that starved period mutants sleep like wild-type flies. By selectively targeting subpopulations of Clk-expressing neurons, we localize the observed sleep phenotype to the dorsally located circadian neurons. These findings show that Clk and cyc act during starvation to modulate the conflict of whether flies sleep or search for food.
Malone*** C, and Brennecke*** J and Dus*** M, Aravin, AA, Sachidanandam, R, Hannon, GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009 May 1; 137(3):522-35. *** authors contributed equally.
In Drosophila gonads, Piwi proteins and associated piRNAs collaborate with additional factors to form a small RNA-based immune system that silences mobile elements. Here, we analyzed nine Drosophila piRNA pathway mutants for their impacts on both small RNA populations and the subcellular localization patterns of Piwi proteins. We find that distinct
piRNA pathways with differing components function in ovarian germ and somatic cells. In the soma, Piwi acts singularly with the conserved flamenco piRNA cluster to enforce silencing of retroviral elements that may propagate by infecting neighboring germ cells. In the germline, silencing programs encoded within piRNA clusters are optimized via a slicer-dependent amplification loop to suppress a broad spectrum of elements. The classes of transposons targeted by germline and somatic piRNA clusters, though not the precise elements, are conserved among Drosophilids, demonstrating that the architecture of piRNA clusters has coevolved with the transposons that they are tasked to control.
Gracheva E, Dus M, Elgin SC. Drosophila RISC component VIG and its homolog Vig2 participate in heterochromatin formation. PLoS One. 2009 Jul 8; 4(7):e6182.
Heterochromatin formation plays an important role in gene regulation and the maintenance of genome integrity. Here we present results from a study of the D. melanogaster gene vig, encoding an RNAi complex component and its homolog vig2 (CG11844) that support their involvement in heterochromatin formation and/or maintenance. Protein null mutations vigEP812 and vig2PL470 act as modifiers of Position Effect Variegation (PEV). VIG and Vig2 are present in polytene chromosomes and partially overlap with HP1. Quantitative immunoblots show depletion of HP1 and HP2 (large isoform) in isolated nuclei from
the vigEP812 mutant. The vig2PL470 mutant strain demonstrates a decreased level of H3K9me2. Pull-down experiments using antibodies specific to HP1 recovered both VIG and Vig2. The association between HP1 and both VIG and Vig2 proteins depends on an RNA component. The above data and the developmental profiles of the two genes suggest that Vig2 may be involved in heterochromatin targeting and establishment early in development, while VIG may have a role in stabilizing HP1/HP2 chromatin binding during later stages.
Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008 Jun 5; 453(7196):798-802.
Drosophila endogenous small RNAs are categorized according to their mechanisms of biogenesis and the Argonaute protein to which they bind. MicroRNAs are a class of ubiquitously expressed RNAs of 22 nucleotides in length, which arise from structured precursors through the action of Drosha–Pasha and Dicer-1– Loquacious complexes1–7. These join Argonaute-1 to regulate gene expression8,9. A second endogenous small RNA class, the Piwiinteracting RNAs, bind Piwi proteins and suppress transposons10,11. Piwi-interacting RNAs are restricted to the gonad, and at least a subset of these arises by Piwi-catalysed cleavage of singlestranded RNAs12,13. Here we show that Drosophila generates a third small RNA class, endogenous small interfering RNAs, in both gonadal and somatic tissues. Production of these RNAs requires Dicer-2, but a subset depends preferentially on
Loquacious1,4,5 rather than the canonical Dicer-2 partner, R2D2 (ref. 14). Endogenous small interfering RNAs arise both from convergent transcription units and from structured genomic loci in a tissue-specific fashion. They predominantly join Argonaute-2 and have the capacity, as a class, to target both protein-coding genes and mobile elements. These observations expand the repertoire of small RNAs in Drosophila, adding a class that blurs distinctions based on known biogenesis mechanisms and functional roles.
Brent, B. and Findley, S.D., Jiang, L., Liu, L., Yin, H., Dus, M, Zhou, P., Elgin, S., and Lin, H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes and Dev. 2007 Sep 15; 21(18):2300-11.
The interface between cellular systems involving small noncoding RNAs and epigenetic change remains largely unexplored in metazoans. RNA-induced silencing systems have the potential to target particular regions of the genome for epigenetic change by locating specific sequences and recruiting chromatin modifiers. Noting that several genes encoding RNA silencing components have been implicated in epigenetic regulation in Drosophila, we sought a direct link between the RNA silencing system and heterochromatin components.
Here we show that PIWI, an ARGONAUTE/PIWI protein family member that binds to Piwi-interacting RNAs (piRNAs), strongly and specifically interacts with heterochromatin protein 1a (HP1a), a central player in heterochromatic gene silencing. The HP1a dimer binds a PxVxL-type motif in the N-terminal domain of PIWI. This motif is required in fruit flies for normal silencing of transgenes embedded in heterochromatin. We also demonstrate that PIWI, like HP1a, is itself a chromatin-associated protein whose distribution in polytene
chromosomes overlaps with HP1a and appears to be RNA dependent. These findings implicate a direct interaction between the PIWI-mediated small RNA mechanism and heterochromatin-forming pathways in determining the epigenetic state of the fly genome.
Brennecke, J., Aravin, A.A., Stark, A., Dus, M, Kellis, M., Sachidanandam, R., and Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007 Mar 23; 128(6):1089-103.
Drosophila Piwi-family proteins have been implicated in transposon control. Here, we examine piwi-interacting RNAs (piRNAs) associated with each Drosophila Piwi protein and find that Piwi and Aubergine bind RNAs that are predominantly antisense to transposons, whereas Ago3 complexes contain predominantly sense piRNAs. As in mammals, the majority of Drosophila piRNAs are derived from discrete genomic loci. These loci comprise mainly defective transposon sequences, and some have previously been identified as master regulators of transposon activity. Our data suggest that heterochromatic piRNA loci interact with potentially active, euchromatic transposons to form an adaptive system for transposon control. Complementary relationships between sense and antisense piRNA populations suggest an amplification loop wherein each piRNA directed cleavage event generates the 50 end of a new piRNA. Thus, sense piRNAs, formed following cleavage of transposon mRNAs may enhance production of antisense piRNAs, complementary to active elements, by directing cleavage of transcripts from master control loci.
Perez-Romero, P. Tyler, R, Abend, JR, Dus, M, and Imperiale, MJ. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J Virol. 2005 Feb 7; 9(4):2366-74.
We previously showed that the adenovirus IVa2 and L1 52/55-kDa proteins interact in infected cells and the IVa2 protein is part of two virus-specific complexes (x and y) formed in vitro with repeated elements of the packaging sequence called the A1-A2 repeats. Here we demonstrate that both the IVa2 and L1 52/55-kDa proteins bind in vivo to the packaging sequence and that each protein-DNA interaction is independent of the other. There is a strong and direct interaction of the IVa2 protein with DNA in vitro. This interaction is observed when probes containing the A1-A2 or A4-A5 repeats are used, but it is not found by using an A5-A6 probe. Furthermore, we show that complex x is likely a heterodimer of IVa2 and an unknown viral protein, while complex y is a monomer or multimer of IVa2. No in vitro interaction of purified L1 52/55-kDa protein with the packaging sequence was found, suggesting that the L1 52/55-kDa protein–DNA interaction may be mediated by an intermediate protein. Results support roles for both the L1 52/55-kDa and IVa2 proteins in
DNA encapsidation.
Li WX, Li H, Lu R, Li F, Dus, M, Atkinson, P, Brydon, EWA., Johnson, KL, García-Sastre, A, Ball, L, Palese, P, and Ding SW. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004 Feb 3; 101(5):1350-5.
Homology-dependent RNA silencing occurs in many eukaryotic cells. We reported recently that nodaviral infection triggers an RNA silencing-based antiviral response (RSAR) in Drosophila, which is capable of a rapid virus clearance in the absence of expression of
a virus-encoded suppressor. Here, we present further evidence to show that the Drosophila RSAR is mediated by the RNA interference (RNAi) pathway, as the viral suppressor of RSAR inhibits experimental RNAi initiated by exogenous double-stranded RNA and RSAR requires the RNAi machinery. We demonstrate that RNAi also functions as a natural antiviral immunity in mosquito cells. We further show that vaccinia virus and human influenza A, B, and C viruses each encode an essential protein that suppresses RSAR in
Drosophila. The vaccinia and influenza viral suppressors, E3L and NS1, are distinct double-stranded RNA-binding proteins and essential for pathogenesis by inhibiting the mammalian IFN-regulated innate antiviral response. We found that the double-stranded RNA-binding domain of NS1, implicated in innate immunity suppression, is both essential and sufficient for RSAR suppression. These findings provide evidence that mammalian virus proteins
can inhibit RNA silencing, implicating this mechanism as a nucleic acid-based antiviral immunity in mammalian cells.
Reviews
Sarangi M and Dus M*. Crème de la Creature: Dietary Influence on Behavior in Animal Models. Frontiers in Behavioral Neuroscience. 29 September 2021.
In this review we highlight how diets high in sugar and fat alter behaviors like eating, learning and memory, courtship and mood; we then consider potential mechanisms and open questions.
Vaziri A and Dus M*. Brain on Food: The Neuroepigenetics of Nutrition. Neurochemistry International. Special feature on Epigenetics. Volume 149, 105099, October 2021.
In this work we review how diet composition affects the brain epigenome and disease risk and discuss future challenges and directions.
May, C and Dus, M*. Confection Confusion: The Interplay between Diet, Taste, and Feeding Behavior. Trends in Endocrinology and Metabolism. Volume 32, Issue 2, Pages 95-105. February 2021.
In this work we review how diets high in added sugar and saturated fats alter taste and the consequences of these changes for feeding behavior and metabolic disease.
Danielle R Reed*, Amber L Alhadeff, Gary K Beauchamp, Nirupa Chaudhari, Valerie B Duffy, Monica Dus, Alfredo Fontanini, John I Glendinning, Barry G Green, Paule V Joseph, George A Kyriazis, Mark Lyte, Padma Maruvada, John P McGann, John T McLaughlin, Timothy H Moran, Claire Murphy, Emily E Noble, M Yanina Pepino, Jennifer L Pluznick, Kristina I Rother, Enrique Saez, Alan C Spector, Catia Sternini, Richard D Mattes. NIH Workshop Report: Sensory Nutrition and Disease, The American Journal of Clinical Nutrition, nqaa302. 09 Dec 2020.
This manuscript highlights the discussion at the NIH NIDDK Panel on the role of sensation in nutrition and disease in November 2019.
Conference Presentations
2022 Gut-Brain Workshop
2022 FENS
2022 ACHEMs
2022 Drosophila Research Conference
2021 Japanese Biochemical Society
2021 EMBL Symposium Metabolism Meets Epigenetics
2021 Annual Meeting of the Japanese Biochemical Society, Epigenetics Symposium
2021 CSHL Neurobiology of Drosophila
2021 European Chemoreceptor Research Organization Symposium, Portugal
2021 Society for the Study of Ingestive Behavior (remote)
2021 Association for Chemoreceptor Sciences Conference (remote)
2020 EMBL Neuroepigenetics Conference, Heidelberg, Germany (remote)
2020 ISOT, Taste and Metabolism panel, Portland, OR (remote)
2020 Association for Chemoreception Sciences
2020 Fusion Neuroepigenetics conference, Bahamas
2019 NIH NIDDK Sensory Science of Nutrition and Disease Symposium
2019 Cold Spring Harbor Neurobiology of Drosophila
2019 European Drosophila Research Conference
2019 Gordon Conference on Neuroethology, Vermont, VT
2019 Society for the Study on Ingestive Behavior, Utrecht, Netherlands
2019 American Chemosensory and Receptor Society Symposium, Florida
2019 National Academy of Science Kavli Frontiers, UC Irvine
2018 GRC Neuromodulation, Switzerland
2018 Helmholtz Diabetes Symposium
2018 MSU Genes and Evolution
2018 CSHL Epigenetics and Chromatin
2018 Fusion Neuroepigenetics Conference
2017 CSHL Neurobiology of Drosophila
2017 Genetics Society of America
2017 ECRO Nutrient Sensing Symposium
2017 Kavli Janelia Conference in Circuits and Behavior
2017 International Society for Endocrinology
2017 American Diabetes Association
2016 EMBL Neurobiology of Obesity
2014 Society for the Study of Ingestive Behavior
2014 Epigenetics and Metabolism

