Keynote speakers
We are delighted to announce our keynote speakers and early career scientist presenters.

Seth Bordenstein
Associate Professor, Departments of Biological Sciences and Pathology, Microbiology, and Immunology, Vanderbilt University
Talk title: The microbiome and Darwin’s mystery of mysteries

Georgiana May
Professor, Department of Ecology, Evolution and Behavior, University of Minnesota
Talk title: Microbial interactions drive the evolution of virulence in pathogens
Presenters

Katherine Amato
Postdoctoral Research Associate, University of Colorado, Boulder
Talk title: Into the wild: exploring the influence of gut microbes on host ecology and behavior
Image credit: Mike Ligouri.

Justine Garcia
Ph.D. Candidate, Emory University
Talk title: Animals in a microbial world: partner fidelity and specificity of horizontal bacterial symbioses in true bugs

Andrea Jani
Research Affiliate, University of Hawaii at Manoa
Talk title: Microbial ecology of an infectious disease: Do symbiotic bacteria protect frogs from the fungal pathogen Batrachochytrium dendrobatidis?

Kevin Kohl
NSF Postdoctoral Research Fellow, Department of Biology, National University of San Luis, Argentina
Talk title: Friends for life: gut microbes allow herbivores to consume toxic plants

Angela Poole
Postdoctoral Research Associate, Departments of Microbiology and Molecular Biology and Genetics, Cornell University
Talk title: Human salivary amylase gene copy number impacts the gut microbiome and its function

Rachel Vannette
Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation, Department of Biology, Stanford University
Talk title: Community assembly and function of the floral microbiome

Kelly Weinersmith
Huxley Fellow, BioSciences Department, Rice University
Talk title: Parasite manipulation of host phenotype: mechanisms, behavior, ecology, and evolution
Keynote speakers
Seth Bordenstein
The microbiome and Darwin’s mystery of mysteries
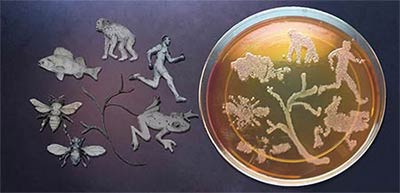 Abstract
Abstract
The life sciences rest on a new footing today in which symbiotic microbes appear essential to nearly every aspect of host form, function, and fitness. As a result, a major research opportunity and challenge is to upgrade general models and experiments of animal and plant evolution in light of the microbiome. In this talk, I will discuss how we are elucidating the varied ways in which the microbiome interacts with the nuclear genome during the origin of new species – what Darwin referred to as the ultimate “mystery of mysteries.” Recent studies of a wide variety of animals have demonstrated that maternal microbial transmission, phylosymbiosis, and hologenomic evolution are worth delving much deeper into.
Georgiana May
Microbial interactions drive the evolution of virulence in pathogens
 Abstract
Abstract
Symbioses are widespread in nature, and the partnering organisms are as diverse as the kingdoms of life. Even as we describe the microbial communities associated with host organisms to increasing depth of “reads,” we lack a depth of understanding in the evolutionary and ecological processes shaping the structure of microbial communities and the symbiotic functions of component species within communities. Here, we seek to understand the role of interactions among symbiont species on the evolution of virulence towards a host. We study a three-way symbiosis of a plant host, an endophytic fungus (commensal with regard to the host), and a plant pathogen. We show that fitness outcomes for the three partners depend on ecological context – the presence/absence of each partner and genotype of interacting partners. We develop a metabolic model for mechanisms underlying differing fitness outcomes by tying together results from metabolic and gene expression analyses. Together, the results show that fitness outcomes are counter-intuitive to classification of organisms at the ends of a spectrum of mutualists and pathogens.
Presenters
Katherine Amato
Into the wild: exploring the influence of gut microbes on host ecology and behavior
 Abstract
Abstract
The gut microbiota is commonly assumed to have co-evolved with mammalian hosts, implying that it positively affects host fitness. However, gut microbiome research often relies on controlled host populations that are not under selective pressure, and it rarely measures host fitness. Data describing variation in the gut microbiota of wild mammals and its impact on host nutrition, physiology, behavior, and ultimately fitness, are critical to broaden our understanding of the gut microbiota and promote new research perspectives. In this talk, I will discuss the impact of host age and sex, seasonal shifts in host diet, and host habitat differences on the gut microbiota of wild, black howler monkeys (Alouatta pigra) in southeastern Mexico. Data suggest that the gut microbiota buffers howler monkeys against nutritional shortfalls during periods of growth and reproduction as well as during periods of reduced energy intake. Relatedly, a reduction in gut microbial diversity appears to contribute to nutritional stress and increased health risks in howler monkeys inhabiting anthropogenically degraded forests. These patterns imply that the gut microbiota has multiple effects on host fitness and provides an impetus for further studies of host-microbe-environment interactions in wild mammals.
Justine Garcia
Animals in a microbial world: partner fidelity and specificity of horizontal bacterial symbioses in true bugs
 Abstract
Abstract
Most animals are colonized by a specific and consistent set of mutualistic microbes. Theory predicts mutualisms persist when both partners benefit, but it is unclear if this is universally true, especially in horizontal symbioses. These symbioses are variable and less likely to be obligate, reducing the incentive to form fair partnerships. How does this affect partner fidelity and symbiont community assembly? Using five true bug species and their bacterial symbionts, I have investigated how partner variation impacts partner specificity and symbiont community assembly. All hosts acquire bacteria in the Burkholderia genus from the environment and store them in crypts, a specialized midgut region. Crypt colonization is consistent with a winnowing model of acquisition as high-throughput sequencing of the midgut indicates the crypts harbor only a subset of the initial bacterial community. Three Burkholderia genotypes are common to all hosts, but many crypts contain multiple genotypes, suggesting common genotypes are the preferred, but not exclusive, constituents of crypt communities. Uncommon symbionts may be less likely to persist within hosts or provide a reduced or alternative benefit. I have tested possible mechanisms that explain the winnowing model, including host immunity and bacterial interactions, and we are currently testing the effect of symbiont genotype on host fitness.
Andrea Jani
Microbial ecology of an infectious disease: Do symbiotic bacteria protect frogs from the fungal pathogen Batrachochytrium dendrobatidis?
 Abstract
Abstract
Symbiotic microbial communities may interact with infectious pathogens sharing a common host. The microbiome may limit pathogen infection or, conversely, an invading pathogen can disturb the microbiome, possibly even leading to secondary disease symptoms due to dysbiosis. Distinguishing between these scenarios is critical to understanding the function and stability of the microbiome. The microbiome-pathogen relationship is of particular interest in the case of Batrachochytrium dendrobatidis , a chytrid fungus that infects the skin of amphibians and has been linked to the decline of a number of amphibian species. I will present coordinated field and laboratory studies that together reveal that B. dendrobatidis infection disturbs and alters the skin‐associated microbiome of the Sierra Nevada yellow‐legged frog (Rana sierrae) during naturally occurring disease outbreaks. Surprisingly, several taxa that might be predicted to inhibit the growth of the pathogen were driven to low abundances by pathogen infection. Furthermore, ongoing preliminary studies of microbiome assembly and stability point to complex interactions between the amphibian host, environment, and chytrid pathogen. I will present the applied implications of these findings for amphibian conservation strategies, and also discuss our results in the broader context of fundamental understanding of the assembly, function, and stability of the microbiome.
Kevin Kohl
Friends for life: gut microbes allow herbivores to consume toxic plants
 Abstract
Abstract
The foraging ecology of mammalian herbivores is strongly shaped by plant secondary compounds (PSCs) that act to defend plants against herbivory. For nearly 40 years it has been hypothesized that gut microbes allow animals to feed on toxic plants. I will present a series of investigations on the gut microbiota of the desert woodrat (Neotoma lepida), some populations of which specialize on highly toxic creosote bush (Larrea tridentata). We demonstrated that the gut microbiota is critical in allowing herbivores to feed on toxic plants. The foregut microbiota exhibited an increased abundance of genes associated with the metabolism of toxic compounds when woodrats were fed creosote PSCs. Treatment with a broad-spectrum antibiotic decreased the ability of woodrats to ingest creosote bush. In addition, natural populations of N. lepida that had no prior experience with creosote bush and are thus naïve to its PSCs, exhibited increased toxin tolerance when inoculated with microbes from experienced individuals. These results demonstrate that microbes can enhance host tolerance to PSCs and thus, potentially expand the dietary niche breadth of wild mammalian herbivores. Furthermore, microbial transfers represent a mechanism by which wild herbivores may rapidly adapt to novel and more potent PSCs brought about by environmental changes.
Angela Poole
Human salivary amylase gene copy number impacts the gut microbiome and its function
 Abstract
Abstract
Humans salivary amylase, encoded by AMY1, hydrolyzes dietary starch into sugars. AMY1 copy number (CN) varies between 1 to 18, and enzyme levels are proportionate to AMY1 CN. Furthermore, human populations with high starch diets have a higher AMY1 CN on average than populations consuming lower amounts of starch, which has been interpreted as evidence of recent selection. Many types of gut microbes also produce amylases as part of short chain fatty acid production. How host AMY1 CN relates to microbiome amylase content and activity is unknown. We addressed this question in a human subjects study involving 25 people selected for either high or low AMY1 CN. Using 16S rRNA gene characterization of microbial community composition, we found that the mean abundances of multiple OTUs, which ferment carbohydrates, were significantly enriched in low AMY1 CN subjects. We transplanted stool collected during the study into germfree mice and assessed their phenotypes after 35 days. Our results showed that mice inoculated with stool from high CN subjects had a higher adiposity than those receiving low CN microbiomes. These results indicate that the copy number of an important dietary gene, AMY1, has an impact on the gut microbiome’s composition and function.
Rachel Vannette
Community assembly and function of the floral microbiome
 Abstract
Abstract
Variation in the process of community assembly can generate communities with divergent structure and function. As a result, predicting and managing the function of the microbiome may benefit from understanding what factors influence its assembly. In this talk, I examine the relative influence of factors that influence community assembly in the floral microbiome, using two different plant species. The results of manipulative laboratory and field experiments, coupled with detailed surveys of bacterial and yeast metacommunities across a landscape, suggest that variation in dispersal, microbial niche axes, and microbe-generated environmental feedbacks, rather than underlying environmental variation in habitat quality or host chemical defense, most strongly influence community composition and nectar chemical characteristics. These results show that understanding the processes by which floral bacterial and yeast communities are assembled can predict their structure and function.
Kelly Weinersmith
Parasite manipulation of host phenotype: mechanisms, behavior, ecology, and evolution
 Abstract
Abstract
Parasites are nature’s hidden ecosystem engineers, as many induce multidimensional changes in their ecosystems (i.e., their hosts) to increase likelihood of transmission. I study the trematode parasite Euhaplorchis californiensis and its California killifish (Fundulus parvipinnis) intermediate host. E. californiensis induces “conspicuous behaviors,” which make the fish up to 30 times more likely to be eaten by the parasite’s definitive host. My research on E. californiensis population ecology suggests that parasites experience higher fitness at high parasite densities, perhaps because parasites share the costs of host manipulation. Scaling up to the impacts of infection on the host ecosystem, I explore how E. californiensis influences host personality and behavioral correlations (i.e., behavioral syndromes), and look for feedbacks between infection and behavior. At the mechanistic level, I examine how E. californiensis manipulates killifish physiology. I found that the interaction between stress intensity and the density of E. californiensis influences stress hormone release rates. Finally, I employ adaptive dynamics modeling to explore how coevolution influences the strength of manipulation, host avoidance behavior and compensation for infection. My research reveals that an ecologically and evolutionarily relevant understanding of host phenotype requires understanding how parasitic ecosystem engineers manipulate the host environment to meet their needs.


