This page outlines the research that’s going on in our lab right now. Background information about our research topic and earlier results can be found in the Introduction and Prior Research Results sections.
Our current research is designed to exploit our recent exciting advances in understanding the molecular basis of cell fate specification. In particular, we are focused on three broad projects:
1. Single-cell RNA Sequencing in Plants
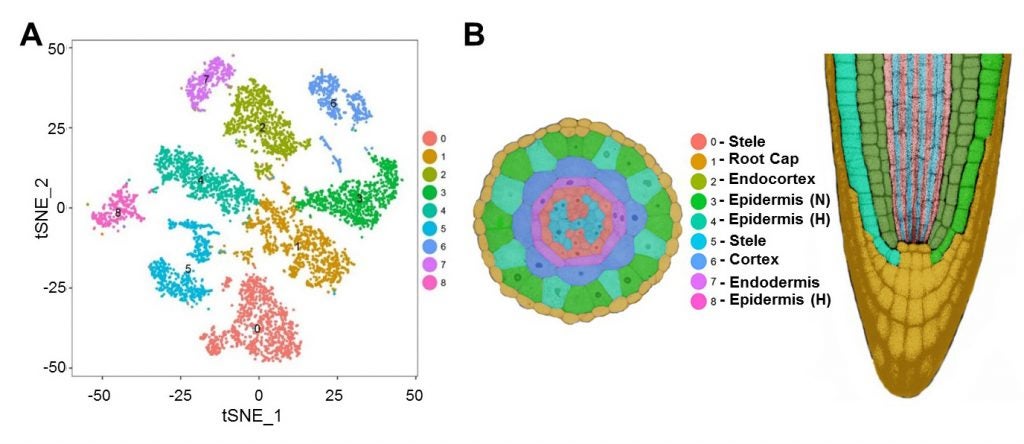
A. Distribution of 7522 individual cell transcriptomes from wild-type Arabidopsis roots into 9 major clusters. B. Assignment of cell clusters to specific root tissues, shown in transverse section (left) and median longitudinal section (right). H=hair cells, N=non-hair cells.
Single-cell RNA sequencing (scRNA-seq) is a relatively new technique for obtaining transcript information from individual cells. Compared to the traditional method of bulk RNA-seq which yields the combined transcript production from a large number of cells, scRNA-seq reveals gene expression from individual cells, providing new insight into cellular heterogeneity, rare cell types, and developmental trajectories. We have successfully adapted a commercially available platform to conduct scRNA-seq on Arabidopsis root cells, and we are continuing to develop and expand the application of scRNA-seq in plants. For more information, read our recent publication (doi: 10.1104/pp.18.01482 [PMID: 30718350]) ) and our protocol for generating root protoplasts for scRNA-seq. Also, these data are available for users to assess the cell-type and developmental-stage expression pattern of any gene of interest…simply visit the Single-Cell eFP Browser at BAR.
2. Root and Root-Hair Development Programs Across the Plant Kingdom
With the availability of many plant genome sequences, it is now possible to analyze and compare gene expression programs across the plant kingdom. We are conducting such a broad comparative transcript profiling study for the developing root and the developing root hairs from a diverse array of plant species. For more information about this project, please see our PGRP project website.
3. Cell-type Pattern Formation in the Root Epidermis.
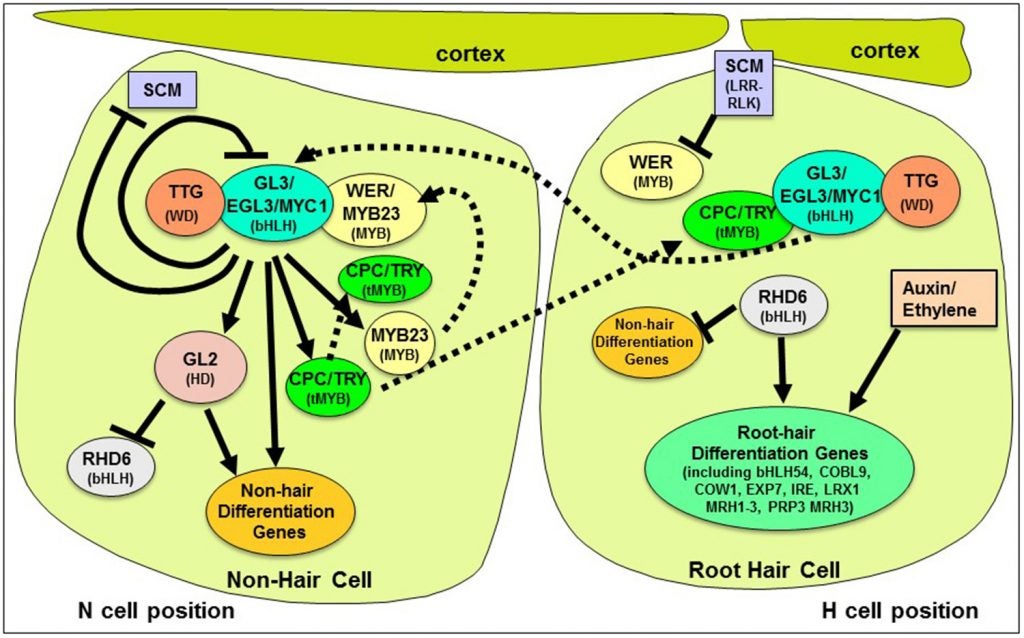
Model for the establishment of root epidermal cell type pattern in Arabidopsis. In brief, an interlocking set of transcriptional feedback loops are influenced by SCM-mediated positional cues to define the expression of the downstream hair and non-hair differentiation genes.
We continue to study the molecular mechanisms that determine the root epidermis cell pattern in Arabidopsis and other species. Here are some of the questions we address:
How do WER and CPC exert opposing effects on cell fate? A central aspect of our model is the proposal that WER and CPC have opposite effects on the cell fate pathway. We are currently employing molecular and biochemical approaches to test three possible explanations for this: (1) CPC directly binds to and inhibits the WER protein, (2) CPC competes with WER by binding to the same promoter element, or (3) CPC competes with WER by binding to the same bHLH protein.
When does this cell fate mechanism begin to act? The mechanism outlined in our model defines the fate of both root and hypocotyl epidermal cells. Because the root and hypocotyl epidermis is derived from a common set of protodermal cells at the globular stage of the Arabidopsis embryo, it is likely that this cell fate mechanism is established during early embryogenesis. We are analyzing the expression of the cell fate genes throughout embryonic development to define the spatial and temporal origin of this patterning mechanism.
What is the evolutionary origin of this patterning mechanism? Arabidopsis and other members of its family (the Brassicaceae family) generates a position-dependent pattern of root epidermal cells, but most plant species outside this family do not display this type of pattern. We are interested in knowing how this cell fate mechanism arose during the evolution of plants. In particular, we are keen to understand the relationship between this cell fate mechanism and alternative mechanisms employed by other plant species.
What are the downstream factors that participate in the larger gene regulatory network? We are using large-scale transcriptomics to define sets of genes that are affected by various mutations and treatments. These studies enable placement of genes and factors relative to one another in an extensive gene regulatory network, and they provoke hypotheses that are being tested using genetic and molecular genetic methods.
Acknowledgements: Our research has been generously supported over the years by the U.S. National Science Foundation, the U.S. Department of Agriculture, and the U.S. Department of Energy.
For more detailed information about our research, please consult our recent publications or contact us directly.

