RNA interference (RNAi) is a recently discovered mechanism of gene regulation that demonstrates an important function of noncoding RNAs. This system utilizes small double-stranded RNAs as a guide to target mRNAs in a sequence specific manner and repress the encoded gene. These double-stranded RNAs are either encoded in the genome as hairpins and termed microRNAs (miRNAs) or originate from exogenous sources such as a virus and termed small-interferring RNAs (siRNAs). The miRNAs or siRNAs are created from longer RNAs by the enzyme Dicer and then loaded with the protein Argonaute to form the RNA-induced silencing complex (RISC) that will lead to repressed translation of a target mRNA. There are still many questions remaining about additional protein players involved in this pathway and the mechanisms of action at each step. We are trying to further understand this system through the following projects:
- Identifying and characterizing components of RISC
- Modeling the kinetics of siRNA and RISC
- Observing miRNA interactions with RISC on a single molecule level
- Single particle tracking of miRNAs in a live cell

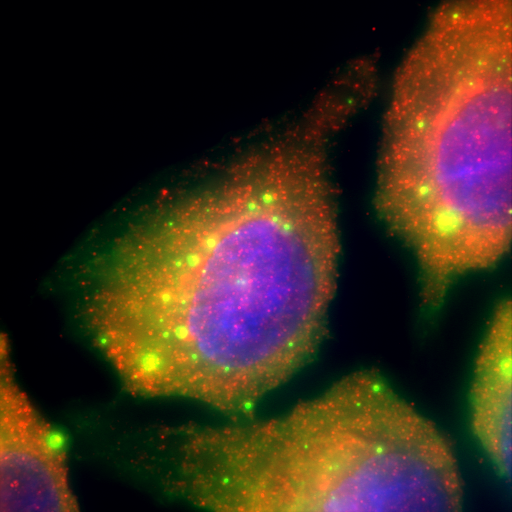
In addition, we have discovered a phenomenon of liquid-liquid phase separation we termed hyperosmotic phase separation (HOPS) that appears widespread with big impact on, e.g., kidney and other stressed cells. We can observe these in live cells and reconstitute them in vitro to study them and their roles as RNA:protein granules.