Membrane transport proteins – channels and energy-coupled pumps – are the molecular gatekeepers of the cell. These proteins import vital nutrients and export dangerous toxins. Microorganisms have contended with different chemical threats throughout evolutionary time, and toxin export is usually the first line of defense . Delving deeply into microbial export systems reveals novel physiologies and previously unknown microbial vulnerabilities. By understanding microorganisms’ intrinsic vulnerabilities, we can generate new leads on antimicrobials and generate basic knowledge about how microbes handle chemical stressors. Our work also provides insight into how bacterial populations have changed and adapted to chemicals and pollutants that humans have introduced to the environment over over the last hundred years, providing a foundation for new bioengineering approaches to mitigate environmental contamination.
Our lab uses a breadth of biochemical and biophysical techniques, including: electrophysiology, membrane protein biochemistry, X-ray crystallography, cryo-EM, and phylogenetic analysis.
Here is a sampling of some of the lab’s current projects:
 Fluoride resistance in the oral microbiome. Microbes associated with oral health resist fluoride using different mechanisms than bacteria associated with dental disease. The figure at left shows fluoride coordination by a bacterial fluoride channel that our lab discovered, Fluc. Our lab is interested in understanding mechanisms of microbial fluoride resistance, and how fluoride modulates the community dynamics of oral biofilms.
Fluoride resistance in the oral microbiome. Microbes associated with oral health resist fluoride using different mechanisms than bacteria associated with dental disease. The figure at left shows fluoride coordination by a bacterial fluoride channel that our lab discovered, Fluc. Our lab is interested in understanding mechanisms of microbial fluoride resistance, and how fluoride modulates the community dynamics of oral biofilms.
Mechanisms of bacterial resistance to household antiseptics, pharmaceutical metabolites, and other chemicals that accumulate in the environment.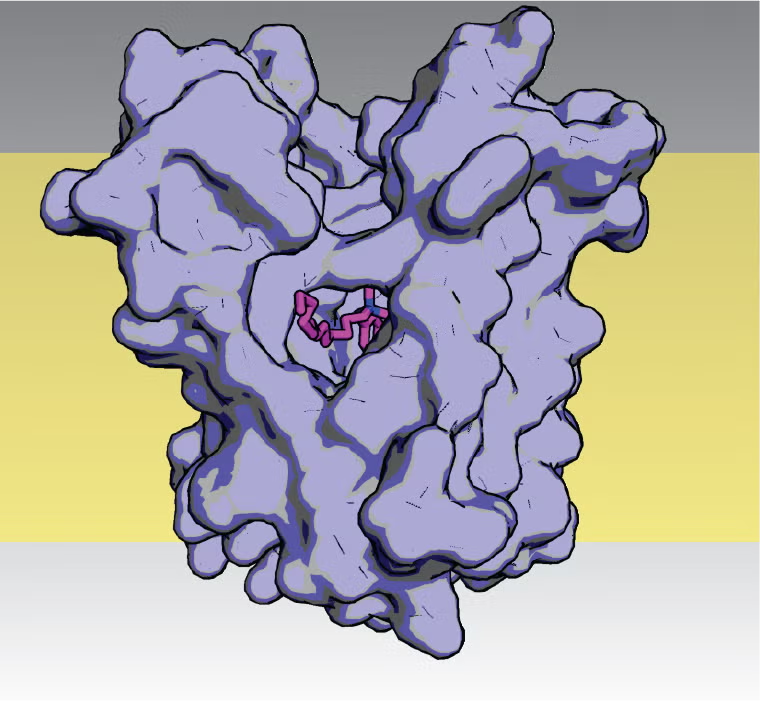 The figure at right shows a model of the E. coli drug exporter from the Small Multidrug Resistance (SMR) protein family in complex with benzalkonium, the active agent in household antiseptics like Lysol. Proteins from the SMR family are responsible for bacterial export of a host of anthropogenic compounds found in wastewater and environmental samples, including antiseptics, agricultural and industrial chemicals, and pharmaceutical byproducts. Our lab uses bioinformatics, crystallography and functional assays to understand the molecular basis of substrate polyspecificity by these proteins.
The figure at right shows a model of the E. coli drug exporter from the Small Multidrug Resistance (SMR) protein family in complex with benzalkonium, the active agent in household antiseptics like Lysol. Proteins from the SMR family are responsible for bacterial export of a host of anthropogenic compounds found in wastewater and environmental samples, including antiseptics, agricultural and industrial chemicals, and pharmaceutical byproducts. Our lab uses bioinformatics, crystallography and functional assays to understand the molecular basis of substrate polyspecificity by these proteins.
Molecular evolution of novel protein folds and functions. We use high throughput mutagenesis, next generation sequencing, and phylogenetics to understand the evolutionary forces that have shaped membrane protein fold and function.
Structural biology approaches for small membrane proteins . Small membrane proteins are notoriously difficult to characterize using crystallography and cryo-EM. Using protein engineering, we are developing new strategies and tools for the structural characterization of small membrane proteins. The panel at right shows a cryo-EM micrograph of 30 kDa membrane protein bound to an antibody fragment.
. Small membrane proteins are notoriously difficult to characterize using crystallography and cryo-EM. Using protein engineering, we are developing new strategies and tools for the structural characterization of small membrane proteins. The panel at right shows a cryo-EM micrograph of 30 kDa membrane protein bound to an antibody fragment.

