
JAMES BARDWELL LAB
RESEARCH
Proteins start life as linear amino acid sequences and end up as beautifully folded, active structures. Dr. Bardwell’s laboratory focuses on recently discovered machinery that drives protein folding in the cell. Powerful genetic, structural, and biophysical tools are being used to generate a detailed picture of how these folding machines work. Members of the Bardwell lab also use directed evolution to improve protein folding. They do this by asking organisms themselves to solve difficult protein-folding problems. By examining the solutions to these problems, they are better able to understand folding in the cell. Our studies include using techniques such as bacterial genetics, biochemistry, biophysics including NMR, X-ray crystallography, and cell biology, microscopy, RNA-seq, RNAi, and CRISPR knockouts

Studying the Physiological Function of Human SERF2
Our lab has successfully discovered the development of amyloid aggregation biosensors in yeast. SERF has been shown to accelerate amyloid aggregation both in vitro and in vivo. It can modulate the disease progression of Alzheimer’s and Parkinson’s Disease in animal models. By determining the physiological functions of SERF, we aim to provide more perspectives on diseases interventions.
SERF2, an RNA G-quadruplex Binding Protein, promotes stress granule formation. Bikash R Sahoo, Xiexiong Deng, Ee Lin Wong, Nathan Clark, Harry Yang, Vivekanandan Subramanian, Bryan B Guzman, Sarah E Harris, Budheswar Dehury, Emi Miyashita, Hirohide Saito, Daniel Dominguez, James CA Bardwell bioRxiv 2023.10.09.561572; doi: https://doi.org/10.1101/2023.10.09.561572
Studying The ATP-Independent Chaperone Spy
Our lab is interested in exploring the functional dynamics in proteins. One area of focus is on the Spy structure. Specifically, how it both allows client folding-while-bound and how it prevents aggregation.
Mitra R, Gadkari VV, Meinen BA, van Mierlo CPM, Ruotolo BT, Bardwell JCA. Mechanism of the small ATP-independent chaperone Spy is substrate specific. Nat Commun. 2021 Feb 8;12(1):851. doi: 10.1038/s41467-021-21120-8

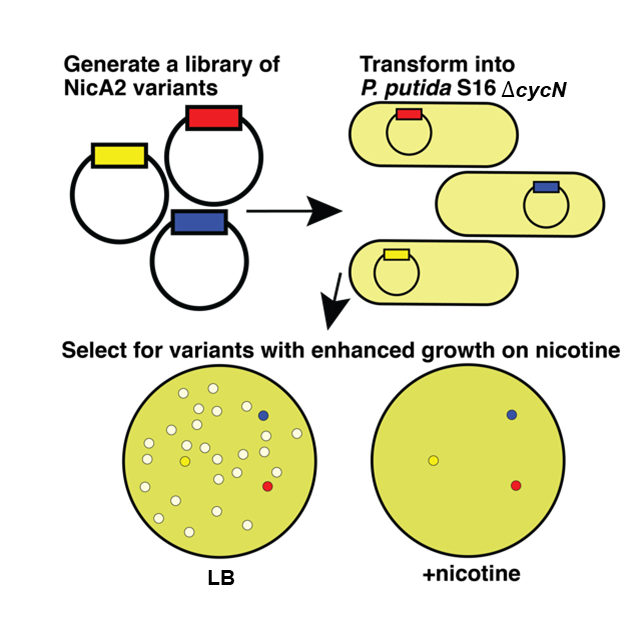
Treating nicotine addiction through directed evolution
Addiction to tobacco is the leading cause of preventable death in the world. We are studying an enzyme NicA2 that degrades nicotine and has been shown to have therapeutic potential when injected into nicotine addicted rats. We find that contrary to the assumption that this enzyme uses oxygen as its electron acceptor, it instead uses a newly discovered cytochrome c, CycN. CycN serves as the natural electron acceptor of NicA2. The inability of CycN mutants to metabolize nicotine in the presence of oxygen opens up the opportunity to select for NicA2 mutants of increased therapeutic potential that can use oxygen directly as an electron acceptor. So far, we have uncovered variants with up to a 200-fold increase in their oxygen-dependent catalytic rate.
Dulchavsky, M., Mitra, R., Wu, K. et al. Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme. Nat Chem Biol (2023). PubMed
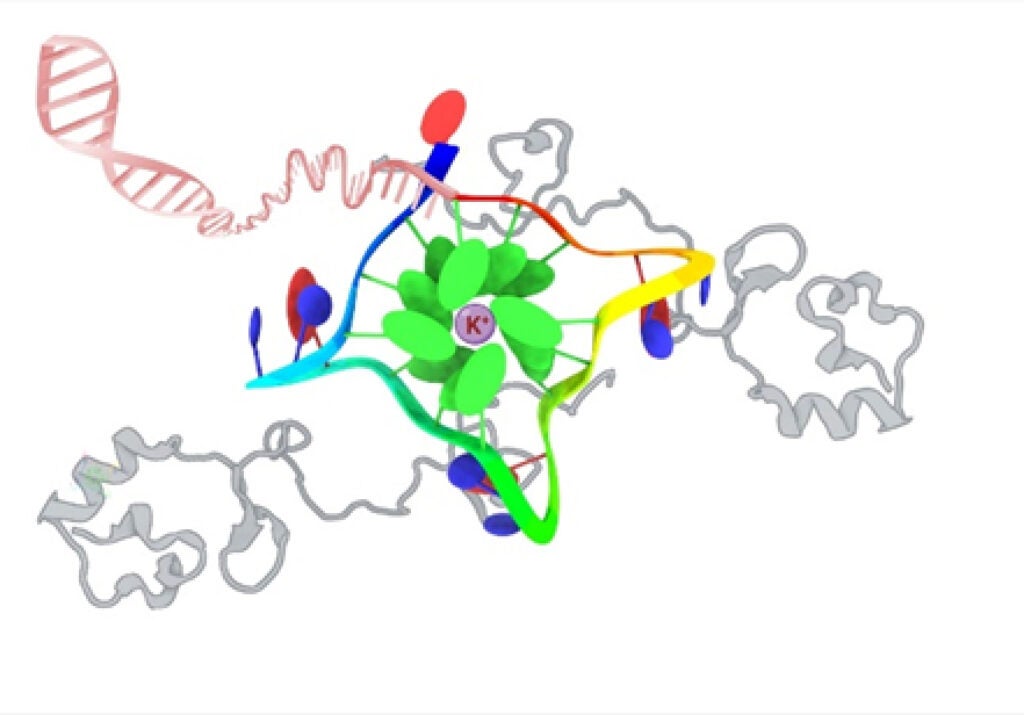
How Proteins Regulate Gene Function By Acting on G-quadruplexes
Our research focuses on elucidating the structural basis of G-quadruplex recognition by two small highly charged and partially disordered proteins. The studied proteins are amenable to solution NMR and preferentially bind to G-quadraplex structures in vitro and in vivo. By understanding how this recognition process occurs we hope to help develop a G-quadruplex binding protein-based platform for gene regulation by manipulating the structure. We approach our studies using biophysical, computational and cellular biology techniques.
Bikash R. Sahoo, Vojč Kocman, Nathan Clark, Nikhil Myers, Xiexiong Deng, Ee L. Wong, Harry J. Yang, Anita Kotar, Bryan B. Guzman, Daniel Dominguez, Janez Plavec, James C.A. Bardwell bioRxiv. Effects of protein G-quadruplex interactions on phase transitions and protein aggregation. 2023.09.21.558871; doi: biorxiv.org

OUR TEAM
James C. Bardwell, Ph.D.
Rowena G. Matthews Collegiate Professor, MCDB HHMI Investigator
- Howard Hughes Medical Investigator
- Department of Molecular Cellular Developmental Biology
- Program in Cellular Molecular Biology
- University of Michigan Biophysics
- Department of Biological Chemistry
Jim received his PhD from the University of Wisconsin in Molecular Biology. He then continued his studies in mRNA Stability at NCI, Bacterial Genetics at Harvard Medical School and Protein Folding at University of Regensburg in Germany. He has has been affiliated with University of Michigan since 1996 and was appointed HHMI investigator in 2005. In Jim’s spare time he enjoys biking, kayaking, and giving tours to Natural History Museum visitors of the coral reef and octopus tanks in his office.
Xiexiong Deng
Postdoctoral Fellow
Xiexiong attained his PhD at Michigan State University in 2018, where he studied the mitotic function of histone H3 in budding yeast. Afterwards, he joined Bardwell lab and studies amyloidogenic protein folding using budding yeast. Outside the lab, he loves watching soccer and playing badminton.
Bikash Sahoo
Postdoctoral Fellow
Bikash received his Ph.D. in Chemistry from Osaka University, Japan, and then joined University of Michigan Biophysics program as a postdoc. Currently, in Bardwell’s lab, he works as a research specialist and his research focuses on the structural biophysics of G-quadruplex binding proteins that regulate genome function and many critical biological processes at high-resolution. Outside the lab, Bikash enjoys designing
websites, 3D illustrations, and browsing international affairs.
Sylvia Widjaja
GraduateLaboratory Technician Student
Sylvia earned her bachelor’s degree of chemical engineering from the University of Melbourne, Australia. She joined the Bardwell-Jakob lab recently as a lab technician to support the research team. She loves spending time with family, traveling and doing outdoor activities.
Harry Yang
Undergraduate
Harry joined the lab in Summer 2021, working with Dr. Xiexiong Deng to understand the physiological function of SERF. He is majoring in Biochemistry and minoring in Computer Science. Outside of lab, he enjoys playing the flute, taking walks, and performing with Revolution Chinese Yoyo.
Matt Crotteau
Undergraduate
Matt joined the lab as a senior in Fall 2022. He is majoring in Molecular, Cellular, and Developmental Biology. He plans to attend graduate school for either Molecular Biology or Bioinformatics after graduation. Matt is from Rice Lake, Wisconsin and enjoys outdoor activities, cooking, and going to the gym. He currently works with graduate student Rishav Mitra on exploring the mechanism of complex coacervation of an intrinsically disordered protein SERF and its interacting RNA along with the mechanistic details of these interactions at a structural level.
Steve Liu
Undergraduate
Steve joined the lab as a sophomore in Fall 2021. He is majoring in Biochemistry and plans to attend graduate school to pursue a PhD. He began his lab work studying under a former graduate student, Mark Dulchavsky, on the Directed Evolution of Flavin-Dependent Nicotine-degradative enzyme (NicA2). Currently, he is working independently on the directed evolution of the other Flavoenzymes. Steve is from Chengdu, China. Outside of the lab, he enjoys cooking, traveling, reading, and painting.
Ken Wan
Senior Laboratory Technician
Ken has worked as a lab technician at U-M for nearly ten years. He joined the Bardwell lab from an HHMI lab in June 2013. Ken’s work is mainly on protein expression and purification. His work consists of making all kinds of constructs for E. coli, Baculovirus insect cell and yeast systems as well as purifying proteins with or without tags using the AKTA system. He enjoys screening and optimizing the protein crystals as well. Ken likes traveling, swimming, and fishing.
Traci Banjanin
Lab Manager & Research Technician II
Traci earned a BS in biochemistry from MSU and a M.Ed. in educational leadership from WSU. After a 30 year career as a chemistry teacher, Traci retired and joined the Bardwell lab in February 2021. She is enjoying supporting the research and administrative needs of all in both the Bardwell and Jakob lab. Outside of work, Traci likes to kayak and boat, bike, read, and cook.
Thea Pazdernik
Undergraduate
Thea joined the lab as a freshman in the Fall of 2022. She is majoring in Molecular, Cellular, and Developmental Biology. She is currently working with undergraduate Steve on the direction evolution of Flavoenzymes. Thea is from Grand Rapids, Michigan, and enjoys traveling, swimming, and spending time with friends and family.
Alejandro Mancilla
Undergraduate
Alejandro joined the lab as a junior in Fall of 2023. He is majoring in Molecular, Cellular, and Developmental Biology. He currently works under Postdoctoral fellow Bikash Sahoo exploring the roles of SERF protein and G-Quadruplex binding on genomic regulation and other important biological processes. Outside of the lab Alejandro enjoys going to the gym, playing soccer, and traveling.
Nathan Clark
Undergraduate
Nathan joined the lab in Winter 2022. He is majoring in Biophysics and minoring in Physics. He currently works with Postdoctoral Fellow Bikash Sahoo to better understand the function of a SERF protein. Nathan is from Hudsonville, Michigan and enjoys hiking, kayaking, reading, and watching movies
Cosmo
Newest
Cosmo joined the Bardwell and Jakob labs in late Oct 2023. He is currently interested in how smells evolve over time. He greatly enjoys meeting new people and long naps. He hopes to follow in his parents pawprints, both of which were flat-coated retriever search and rescue dogs.
Lizzie Kinsey
Lizzie joined the lab in the Fall of 2023. She is majoring in Ecology, Evolution, and Biodiversity and minoring in Spanish. Currently, she works with Dr. Bardwell and Carl Pate to study the maintenance and regeneration of the Clytia Hemisphaerica. Outside the lab, she enjoys swimming, painting, and spending time at the beach..
Tejas Navaratna
Tejas got his PhD from the University of Michigan in 2020 and is excited to be back working on enzyme discovery through natural and directed evolution, as well as applying reaction modeling and spectroscopy to understand mechanism. He is from San Jose, California and enjoys the outdoors and good coffee.
Carl Pate
Carl joined the lab as a freshman in 2023 working with Dr. Bardwell on the cultivation of Clytia jellyfish for regeneration research. He is an undergraduate student studying mechanical engineering and an avid reef tank enthusiast having 2 systems of his own. Outside the lab he participates on the ATLO sub-team of MASA, plays the tuba in the University and Campus band, and is an Eagle Scout.
Ammar Ali
Ammar joined the lab as a junior in the Winter of 2024. He is majoring in Molecular, Cellular, and Developmental Biology. He works with Postdoctoral fellow Bikash Sahoo studying the structural biophysics of the G-quadruplexes in relation to the SERF2 and other associated proteins. Outside of the lab, Ammar loves to watch and play sports with friends, workout, spend time with family, and volunteer at the children’s hospital.
SELECTED PUBLICATIONS

Effects of protein G-quadruplex interactions on phase transitions and protein aggregation
Bikash R. Sahoo, Vojč Kocman, Nathan Clark, Nikhil Myers, Xiexiong Deng, Ee L. Wong, Harry J. Yang, Anita Kotar, Bryan B. Guzman, Daniel Dominguez, Janez Plavec, James C.A. Bardwell. Effects of protein G-quadruplex interactions on phase transitions and protein aggregation. bioRxiv 2023.09.21.558871; doi: biorxiv.org
We show that human protein Znf706 interacts specifically with stable, non-canonical nucleic acid structures known as G-quadruplexes. Znf706, though only 76 residues long, is comprised of two distinct domains, one dis-ordered and one ordered. The disordered domain is homologous to the SERF family of proteins and acts to accelerate amyloid formation. The ordered domain contains a C2H2 type zinc-finger. We show that Znf706 not only accelerates amyloid formation but also accelerates amorphous protein aggregation. We find that Znf706 binds preferentially to parallel G-quadruplexes…Read More
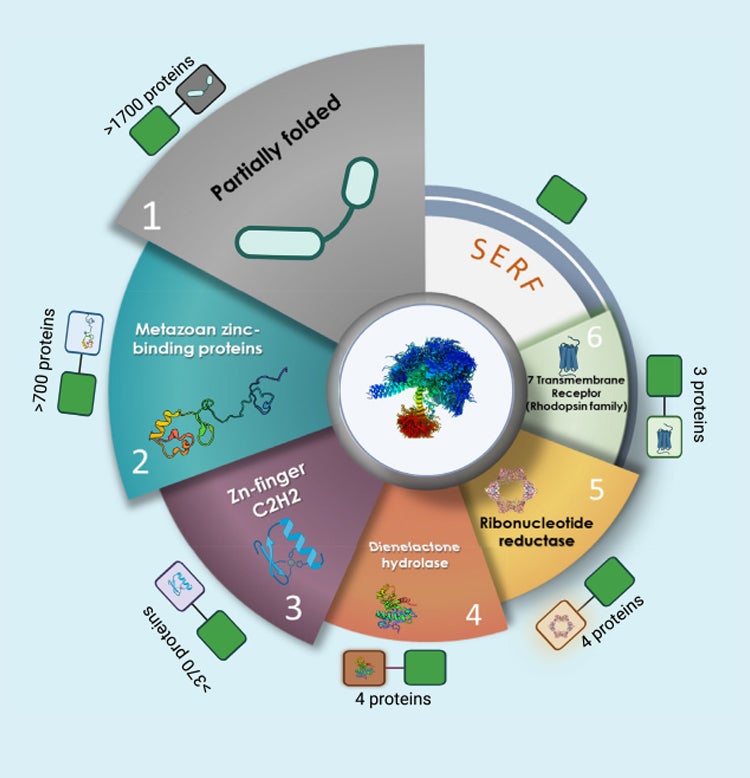
SERF, a family of tiny highly conserved, highly charged proteins with enigmatic functions
Sahoo BR, Bardwell JCA. SERF, A Family Of Tiny Highly Conserved, Highly Charged Proteins With Enigmatic Functions. The FEBS Journal. 2022 Jun 13doi: 10.0000/febs,16555. PubMed
Amyloid formation is a misfolding process that has been linked to age-related diseases, including Alzheimer’s and Huntington’s. Understanding how cellular factors affect this process in vivo is vital in realizing the dream of controlling this insidious process that robs so many people of their humanity. SERF (small EDRK-rich factor) was initially isolated as a factor that accelerated polyglutamine amyloid formation in a C…Read More

Mechanism of the small ATP-independent chaperone Spy is substrate specific
Mitra R, Gadkari VV, Meinen BA, van Mierlo CPM, Ruotolo BT, Bardwell JCA. Mechanism of the small ATP-independent chaperone Spy is substrate specific. Nat Commun. 2021 Feb 8;12(1):851. doi: 10.1038/s41467-021-21120-8. PubMed
ATP-independent chaperones are usually considered to be holdases that rapidly bind to non-native states of substrate proteins and prevent their aggregation. These chaperones are thought to release their substrate proteins prior to their folding. Spy is an ATP-independent chaperone that acts as an aggregation inhibiting holdase but does so by allowing its substrate proteins to fold while they remain continuously chaperone bound, thus acting as a foldase as well…Read More

Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme
Dulchavsky, M., Mitra, R., Wu, K. et al. Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme. Nat Chem Biol (2023). Nature.com
The flavoenzyme nicotine oxidoreductase (NicA2) is a promising injectable treatment to aid in the cessation of smoking, a behavior responsible for one in ten deaths worldwide. NicA2 acts by degrading nicotine in the bloodstream before it reaches the brain. Clinical use of NicA2 is limited by its poor catalytic activity in the absence of its natural electron acceptor CycN. Without CycN, NicA2 is instead oxidized slowly by dioxygen (O2), necessitating unfeasibly large doses in a therapeutic setting. Here, we report a genetic selection strategy that directly links CycN-independent activity of NicA2 to growth of Pseudomonas putida S16. This selection enabled us to evolve NicA2 variants with substantial improvement in their rate of oxidation by O2. The encoded mutations cluster around a putative O2 tunnel, increasing flexibility and accessibility to O2 in this region. These mutations further confer desirable clinical properties. A variant form of NicA2 is tenfold more effective than the wild type at degrading nicotine in the bloodstream of rats…Read More

A metabolite binding protein moonlights as a bile-responsive chaperone
Lee C, Betschinger P, Wu K, Zyla DS, Glockshuber R, Bardwell JCA. A metabolite binding protein moonlights as a bile-responsive chaperone. EMBO J. 2020 Sep 3;e104231. doi: 10.15252/embj.2019104231. Online ahead of print. PubMed
Bile salts are secreted into the gastrointestinal tract to aid in the absorption of lipids. In addition, bile salts show potent antimicrobial activity in part by mediating bacterial protein unfolding and aggregation. Here, using a protein folding sensor, we made the surprising discovery that the Escherichia coli periplasmic glycerol-3-phosphate (G3P)-binding protein UgpB can serve, in the absence of its substrate, as a potent molecular chaperone that exhibits anti-aggregation activity against bile salt-induced protein aggregation…Read More

SERF engages in a fuzzy complex that accelerates primary nucleation of amyloid proteins
Meinen BA, Gadkari VV, Stull F, Ruotolo BT, Bardwell JCA. SERF engages in a fuzzy complex that accelerates primary nucleation of amyloid proteins. Proc Natl Acad Sci USA 2019 Oct 28. pii: 201913316. doi: 10.1073/pnas.1913316116. PubMed [FACULTY OPINIONS]
The assembly of small disordered proteins into highly ordered amyloid fibrils in Alzheimer’s and Parkinson’s patients is closely associated with dementia and neurodegeneration. Understanding the process of amyloid formation is thus crucial in the development of effective treatments for these devastating neurodegenerative diseases. Recently, a tiny, highly conserved and disordered protein called SERF was discovered to modify amyloid formation in Caenorhabditis elegans and humans…Read More

In vivo chloride concentrations surge to proteotoxic levels during acid stress
Stull F, Hipp H, Stockbridge RB, Bardwell JCA. In vivo chloride concentrations surge to proteotoxic levels during acid stress. Nature Chemical Biology 2018 Oct; 14, 1051-1058. PubMed This paper was the subject of a News and Views [PDF] citation in Nature Chemical Biology by Colin Kleanthous. [FACULTY OPINIONS]
To successfully colonize the intestine, bacteria must survive passage through the stomach. The permeability of the outer membrane renders the periplasm of Gram-negative bacteria vulnerable to stomach acid, which inactivates proteins. Here we report that the semipermeable nature of the outer membrane allows the development of a strong Donnan equilibrium across this barrier at low pH. As a result, when bacteria are exposed to conditions that mimic gastric juice, periplasmic chloride concentrations rise to levels that exceed 0.6 M…Read More
Determining crystal structures through crowdsourcing and coursework
Horowitz S, Koepnick B, Martin R, Tymieniecki A, Winburn AA, Cooper S, Flatten J, Rogawski DS, Koropatkin NM, Hailu TT, Jain N, Koldewey P, Ahlstrom LS, Chapman MR, Sikkema AP, Skiba MA, Maloney FP, Beinlich FR; Foldit Players; University of Michigan students, Popović Z, Baker D, Khatib F, Bardwell JC. (2016). Determining crystal structures through crowdsourcing and coursework. Nat. Commun.2016 Sep 16;7:12549. doi: 10.1038/ncomms12549. PubMed
This paper was the subject of a Marketplace Tech interview.
We show here that computer game players can build high-quality crystal structures. Introduction of a new feature into the computer game Foldit allows players to build and real-space refine structures into electron density maps…Read More

Visualizing chaperone-assisted protein folding
Horowitz S, Salmon L, Koldewey P, Ahlstrom L, Martin R, Quan S, Afonine PV, van den Bedem H, Wang L, Xu Q, Trievel RC, Brooks CL, Bardwell JCA. (2016) Visualizing Chaperone-assisted Folding. Nature Struct. Mol. Biol. 2016 Jul;23(7):691-7. doi: 10.1038/nsmb.3237. Epub 2016 May 30. PubMed This paper was the subject of a News and Views citation in NSMB by Patricia L. Clark and Adrian H. Elcock.
Challenges in determining the structures of heterogeneous and dynamic protein complexes have greatly hampered past efforts to obtain a mechanistic understanding of many important biological processes. One such process is chaperone-assisted protein folding. Obtaining structural ensembles of chaperone-substrate complexes would ultimately reveal how chaperones help proteins fold into their native state. To address this problem, we devised a new structural biology approach based on X-ray crystallography, termed residual electron and anomalous density (READ)…Read More

Forces Driving Chaperone Action
Koldewey P, Stull F, Horowitz S, Martin R, Bardwell JCA. (2016) Forces Driving Chaperone Action. Cell. 2016 Jun 9. pii: S0092-8674(16)30657-2. doi: 10.1016/j.cell.2016.05.054. PubMed This paper was the subject of a News and Views [PDF] citation in NSMB by Patricia L. Clark and Adrian H. Elcock.
It is still unclear what molecular forces drive chaperone-mediated protein folding. Here, we obtain a detailed mechanistic understanding of the forces that dictate the four key steps of chaperone-client interaction: initial binding, complex stabilization, folding, and release. Contrary to the common belief that chaperones recognize unfolding intermediates by their hydrophobic nature..Read More

Chaperone activation by unfolding
Foit L, George JS, Zhang BW, Brooks CL 3rd, Bardwell JC. (2013) Chaperone activation by unfolding. Proc Natl Acad Sci U S A. 110(14):E1254-62. Epub 2013 Mar 4. PMCID:PMC3619340. PubMed This paper was the subject of a Commentary [PDF] by Karan S. Hingorani and Lila M. Gierasch.
Conditionally disordered proteins can alternate between highly ordered and less ordered configurations under physiological conditions. Whereas protein function is often associated with the ordered conformation, for some of these conditionally unstructured proteins, the opposite applies: Their activation is associated with their unfolding. An example is the small periplasmic chaperone HdeA, which is critical for the ability of enteric bacterial pathogens like Escherichia coli to survive passage through extremely acidic environments, such as the human stomach. At neutral pH, HdeA is a chaperone-inactive dimer. On a shift to low pH, however, HdeA monomerizes…Read More
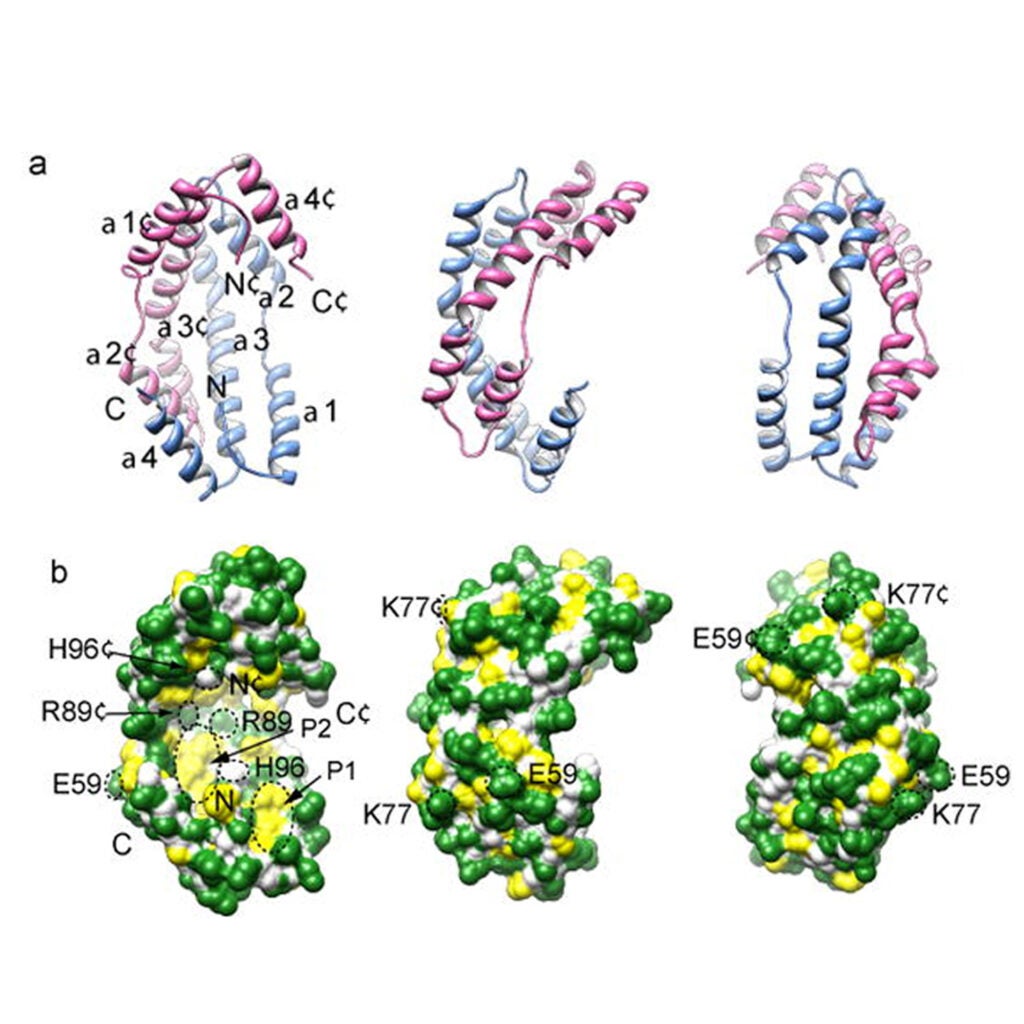
Genetic selection designed to stabilize proteins uncovers a chaperone called Spy
Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JC. (2011) Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol. 18(3):262-9. PubMed This paper was the subject of a “News and Views” citation in Nature by Evan T. Powers and Willian E. Balch.
To optimize the in vivo folding of proteins, we linked protein stability to antibiotic resistance, thereby forcing bacteria to effectively fold and stabilize proteins…Read More

Optimizing protein stability in vivo
Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JC. (2009) Optimizing protein stability in vivo. Mol Cell. Dec 11;36(5):861-71. PubMed
Bile salts are secreted into the gastrointestinal tract to aid in the absorption of lipids. In addition, bile salts show potent antimicrobial activity in part by mediating bacterial protein unfolding and aggregation. Here, using a protein folding sensor, we made the surprising discovery that the Escherichia coli periplasmic glycerol-3-phosphate (G3P)-binding protein UgpB can serve, in the absence of its substrate, as a potent molecular chaperone that exhibits anti-aggregation activity against bile salt-induced protein aggregation…Read More
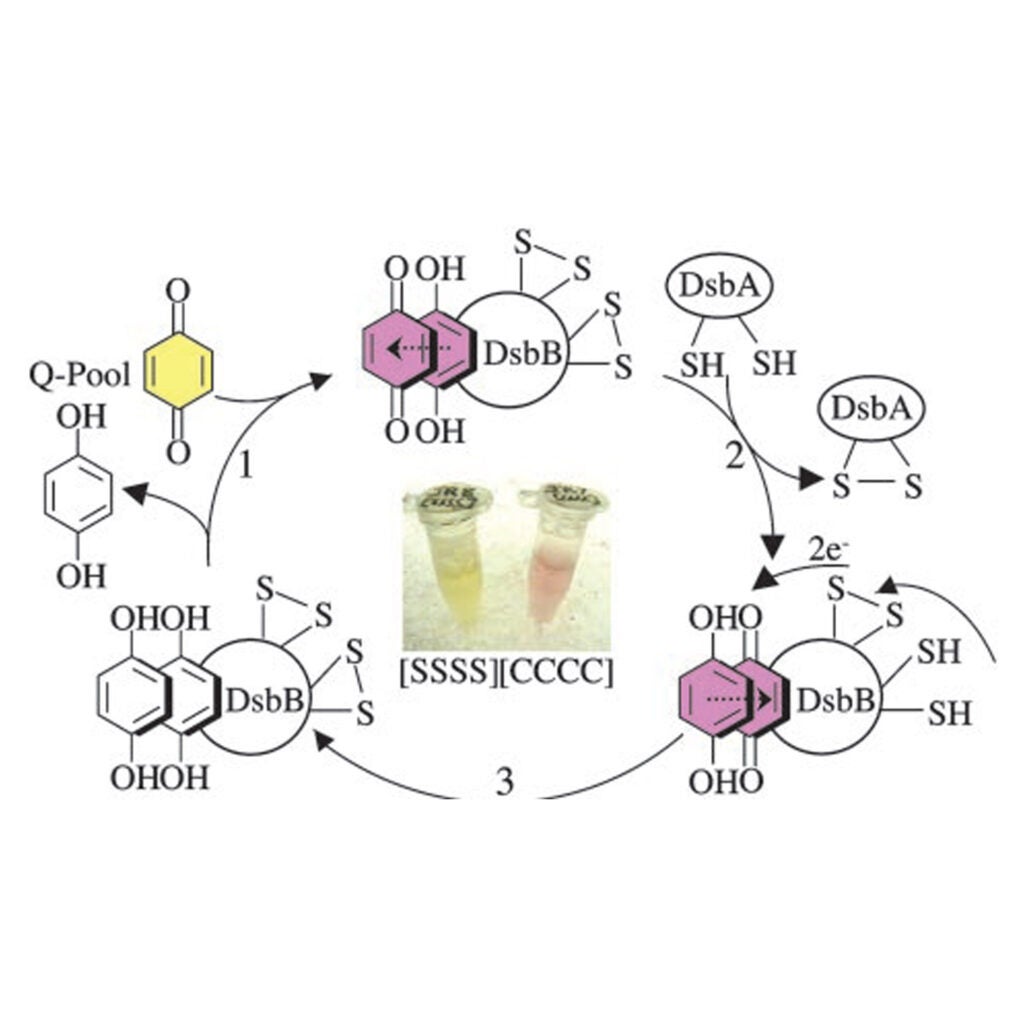
Disulfide bond formation involves a quinhydrone-type charge–transfer complex
Regeimbal J, Gleiter S, Trumpower BL, Yu CA, Diwakar M, Ballou DP, Bardwell JC. (2003) Disulfide bond formation involves a quinhydrone-type charge-transfer complex. Proc Natl Acad Sci. 2003 Nov 25; 100(24):13779-84.

Protein oxidation: prime suspect found ‘not guilty’
Bader M, Winther JR, and Bardwell JC. (1999) Protein oxidation: prime suspect found ‘not guilty’. Nat Cell Biol. Jul;1(3):E57-8. PubMed
Glutathione has long been suspected to be the primary source of oxidative power for protein
folding. It has now been shown to be just the opposite, namely a source of reductants. The ultimate
origin of oxidants has become even more of a mystery…Read More

Chaperone activity with a redox switch
Jakob U, Muse W, Eser M, and Bardwell JC. (1999) Chaperone activity with a redox switch Cell Feb 5;96(3):341-52. PubMed
Hsp33, a member of a newly discovered heat shock protein family, was found to be a very potent molecular chaperone. Hsp33 is distinguished from all other known molecular chaperones by its mode of functional regulation. Its activity is redox regulated. ..Read More

A pathway for disulfide bond formation in vivo
Bardwell JC, Lee JO, Jander G, Martin N, Belin D, and Beckwith J. (1993) A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci Feb 1;90(3):1038-42. PubMed
Protein disulfide bond formation in Escherichia coli requires the periplasmic protein DsbA. We describe here mutations in the gene for a second protein, DsbB, which is also necessary for disulfide bond formation. Evidence suggests that DsbB may act by reoxidizing DsbA, thereby regenerating its ability to donate its disulfide bond to target proteins. We propose that DsbB, an integral membrane protein, may be involved in transducing redox potential across the cytoplasmic membrane…Read More
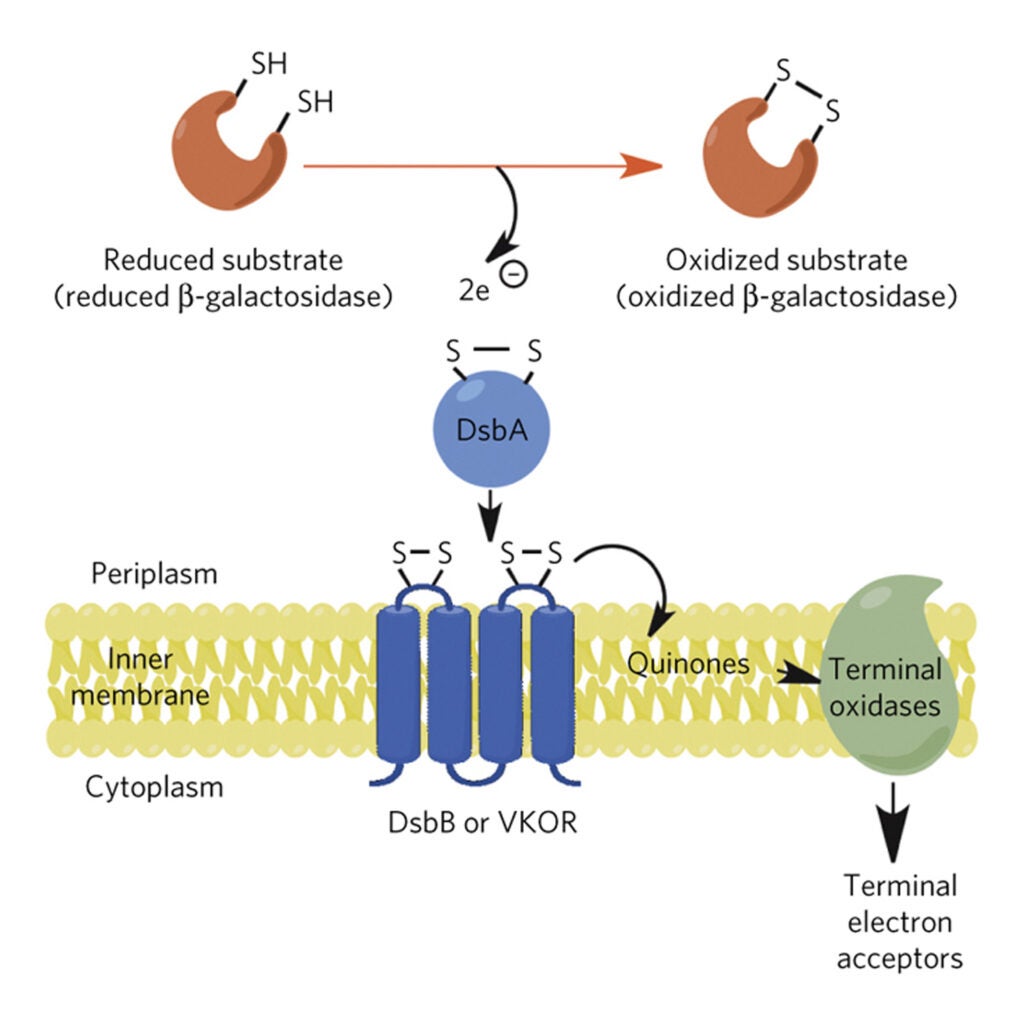
Identification of a protein required for disulfide bond formation in vivo
Bardwell JC, McGovern K, and Beckwith J. (1991) Identification of a protein required for disulfide bond formation in vivo. Cell Nov 1;67(3):581-9. PDF
We describe a mutation (dsbA) that renders Escherichia coli severely defective in disulfide bond formation. In dsbA mutant cells, pulse-labeled beta-lactamase, alkaline phosphatase, and OmpA are secreted but largely lack disulfide bonds. These disulfideless proteins may represent in vivo folding intermediates, since they are protease sensitive and chase slowly into stable oxidized forms. The dsbA gene codes for a 21,000 Mr periplasmic protein containing the sequence cys-pro-his-cys, which resembles the active sites of certain disulfide oxidoreductases. The purified DsbA protein is capable of reducing the disulfide bonds of insulin, an activity that it shares with these disulfide oxidoreductases…Read More

JOIN OUR LAB
If you are interested in joining a well-funded, collaborative team working in the areas of protein structure, function and genetic engineering, send Jim your CV via email to: Jbardwel@umich.edu. We are recruiting for postdocs, visiting students (both domestic and foreign) and graduate students. To see how successful our visiting students and scholars have been click here.
Connect with our Instagram





CONTACT
Location
HHMI @ The University of Michigan Biological Sciences Building 1105 N University , Room 5022 Ann Arbor, Michigan 48109






















