Keynote Speakers
 Steven A. Frank
Steven A. Frank
Professor, Ecology and Evolutionary Biology, University of California, Irvine
Three unsolved puzzles in infectious disease
 Unsolved puzzles provide a window onto the limits of current understanding and opportunities for new work. I describe three unsolved puzzles of infectious disease. First, the number of bacterial cells (dosage) required to start an infection varies over several orders of magnitude for different pathogens. Second, influenza evolves rapidly to escape host immunity, whereas measles does not evolve escape variants. Third, humans frequently exposed to zoonotic pathogens in animals seem often to develop immunity without overt symptoms, whereas those exposed sporadically appear to develop more severe disease. Following these puzzles leads to fundamental aspects of molecular pathogenesis, immunity, route of infection, and evolutionary dynamics.
Unsolved puzzles provide a window onto the limits of current understanding and opportunities for new work. I describe three unsolved puzzles of infectious disease. First, the number of bacterial cells (dosage) required to start an infection varies over several orders of magnitude for different pathogens. Second, influenza evolves rapidly to escape host immunity, whereas measles does not evolve escape variants. Third, humans frequently exposed to zoonotic pathogens in animals seem often to develop immunity without overt symptoms, whereas those exposed sporadically appear to develop more severe disease. Following these puzzles leads to fundamental aspects of molecular pathogenesis, immunity, route of infection, and evolutionary dynamics.
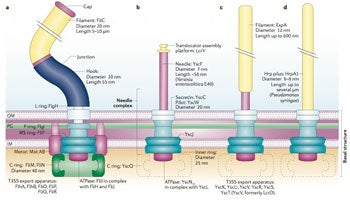
Image: basal structure, Nature Reviews Microbiology
 Caroline Buckee
Caroline Buckee
Assistant Professor of Epidemiology
Department of Epidemiology, Harvard School of Public Health
The effects of cross-immunity and competition on meningococcal evolution
The meningococcus, Neisseria meningitidis, is a human commensal bacterium that occasionally causes invasive disease. Meningococcal populations are characterized by structured diversity, with particular combinations of both antigenic determinants and housekeeping loci persisting across wide geographical and long temporal scales despite evidence for extensive genetic exchange between lineages. The evolutionary mechanisms maintaining this structure in the face of continual diversity generation remain unclear.
Over the past several years we have analyzed the population structures of several antigenic loci and housekeeping genes from a number of large meningococcal carriage data sets, and developed theoretical frameworks and data analysis tools to identify possible evolutionary mechanisms shaping them. I will present these meningococcal data sets, first putting the meningococcal population structure in the context of the range of bacterial population structures that exist among human pathogens. The development of models exploring the effects of cross-immunity and within-host competition on pathogen population structure will then be presented, as well as the insights gained from them into the impact of stochastic processes and spatial structure on bacterial populations. Lastly, the challenges of calibrating meningococcal data sets with model outcomes will be discussed.
 Silvie Huijben
Silvie Huijben
Postdoctoral Scholar, Penn State, Center for Infectious Disease Dynamics
The ecology and evolution of resistant malaria parasites
 The evolution of drug resistance in malaria parasites is to a large extent determined by the within-host ecology of resistant and sensitive parasites. Human malaria infections frequently consist of more than one genotype; a better understanding of the ecology and evolution of malaria parasites will facilitate evolutionary-informed rational treatment strategies that reduce selection for resistance.
The evolution of drug resistance in malaria parasites is to a large extent determined by the within-host ecology of resistant and sensitive parasites. Human malaria infections frequently consist of more than one genotype; a better understanding of the ecology and evolution of malaria parasites will facilitate evolutionary-informed rational treatment strategies that reduce selection for resistance.
Using the well-established rodent malaria model Plasmodium chabaudi, I studied the fitness of resistant parasites in the following ecological contexts: (i) in multi-genotype infections, (ii) at different genotype frequencies and (iii) under varying drug pressures. Selection coefficient analysis, derived from ecological statistics, was introduced to measure the strength and direction of selection on resistant parasites. In addition to studying competition between asexual parasite stages at the within-host level, sexual transmission stages were studied to understand how these ecological dynamics could play out at the between-host level.
The fitness of resistant parasites strongly depended on ecological context, but not always in intuitive ways. Altering the within-host ecology by administrating drugs invariably resulted in strong positive selection for drug-resistant parasites. Reduction of drug pressure led to significant decreases in selection for resistance, while being equally as effective, or even better, at improving host health and reducing infectiousness.
Photo: Brian Chan
 David Kennedy
David Kennedy
Ph.D. Candidate, Department of Ecology and Evolution, University of Chicago
The role of multilevel selection in the maintenance of viral genetic diversity
 The next step in host-pathogen evolution studies is to test models that incorporate both within-host and between-host dynamics. These models are often difficult to apply, however, because very little is known about within-host parameters. Furthermore, the relationship between genotypic variability and phenotypic variability in most virus systems is poorly characterized. The gypsy moth caterpillar and its associated baculovirus provide us with an opportunity to solve both of these problems. To learn about the pathogen processes occurring within a caterpillar, I fit a suite probabilistic birth-death models to dose-response-time data, by using a novel MCMC algorithm that combines parameter line-searches, principal component analysis and MCMC. I then set up a probabilistic between-host model that incorporates a within-host birth-death model, thus allowing for both mechanism-based response-times and within-host competition. Model and data together suggest a tradeoff between within-host competition and between-host competition. This result is supported by phenotypic data and preliminary genotypic data that show high levels of polymorphism in the baculovirus population. Currently, I am attempting to expand the model to allow for recombination between virus strains by creating and incorporating a phenotype-genotype map. This will provide a rare opportunity to explore how selection, recombination and drift shape a pathogen population.
The next step in host-pathogen evolution studies is to test models that incorporate both within-host and between-host dynamics. These models are often difficult to apply, however, because very little is known about within-host parameters. Furthermore, the relationship between genotypic variability and phenotypic variability in most virus systems is poorly characterized. The gypsy moth caterpillar and its associated baculovirus provide us with an opportunity to solve both of these problems. To learn about the pathogen processes occurring within a caterpillar, I fit a suite probabilistic birth-death models to dose-response-time data, by using a novel MCMC algorithm that combines parameter line-searches, principal component analysis and MCMC. I then set up a probabilistic between-host model that incorporates a within-host birth-death model, thus allowing for both mechanism-based response-times and within-host competition. Model and data together suggest a tradeoff between within-host competition and between-host competition. This result is supported by phenotypic data and preliminary genotypic data that show high levels of polymorphism in the baculovirus population. Currently, I am attempting to expand the model to allow for recombination between virus strains by creating and incorporating a phenotype-genotype map. This will provide a rare opportunity to explore how selection, recombination and drift shape a pathogen population.
Photo: top right – Libby Eakin, leaf – Alison Hunter
 Britt Koskella
Britt Koskella
Postdoctoral Fellow, Department of Zoology, University of Oxford
Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria
 It is increasingly recognized that the ecological and evolutionary consequences of host-parasite interactions are shaped by the spatial scale at which parasites adapt to hosts. For hyperparasite-parasite interactions, coevolution is likely to be influenced by the host of the parasite and to differ among within-host environments. In this talk I will examine the scale at which bacteriophages adapt to infect their host bacteria within natural populations living in and on their plant host, the horse chestnut tree. This will include data suggesting that phages are targeting common bacterial species, including an important plant pathogen, within plant host tissues and that the scale of bacteria-phage interactions is shaped primarily by the tree host rather than geographic distance; phages are more infective to bacteria from the same tree, relative to other trees, but do not differ in infectivity on bacteria from different leaves within the same tree. The results reinforce the developing idea that selection mosaics should be common across multitrophic interactions. I discuss these findings both in light of phage therapy for regulating bacterial populations and, more generally, to highlight the importance of understanding the spatial scale and biotic complexity of species interactions in successfully predicting the outcome of coevolution.
It is increasingly recognized that the ecological and evolutionary consequences of host-parasite interactions are shaped by the spatial scale at which parasites adapt to hosts. For hyperparasite-parasite interactions, coevolution is likely to be influenced by the host of the parasite and to differ among within-host environments. In this talk I will examine the scale at which bacteriophages adapt to infect their host bacteria within natural populations living in and on their plant host, the horse chestnut tree. This will include data suggesting that phages are targeting common bacterial species, including an important plant pathogen, within plant host tissues and that the scale of bacteria-phage interactions is shaped primarily by the tree host rather than geographic distance; phages are more infective to bacteria from the same tree, relative to other trees, but do not differ in infectivity on bacteria from different leaves within the same tree. The results reinforce the developing idea that selection mosaics should be common across multitrophic interactions. I discuss these findings both in light of phage therapy for regulating bacterial populations and, more generally, to highlight the importance of understanding the spatial scale and biotic complexity of species interactions in successfully predicting the outcome of coevolution.
 Thierry Lefevre
Thierry Lefevre
Postdoctoral Fellow, Biology Department, Emory University
The role of host behavior in host-parasite interactions and disease dynamics
 Host-parasite interactions are often characterized by antagonistic coevolutionary processes: parasites evolve to optimize their host exploitation and transmission, while hosts evolve to minimize parasite-induced fitness losses. This arms race can result in fascinating adaptations in both hosts and parasites. For instance, there can be parasite manipulation of host behavior whereby parasites alter the behavior of their host in a way that increases their own transmission. In contrast, hosts may engage in self-medication, a series of behaviors through which animals exploit the anti-parasitic properties of a third species to prevent or reduce infection. Parasitic manipulation and self-medication both lead to substantial host behavioral changes with contrasting consequences on disease dynamics. Here, I will present some experimental results on parasitic manipulation in vector-borne diseases and on self-medication in monarch butterflies and will discuss their potential effects on disease transmissions.
Host-parasite interactions are often characterized by antagonistic coevolutionary processes: parasites evolve to optimize their host exploitation and transmission, while hosts evolve to minimize parasite-induced fitness losses. This arms race can result in fascinating adaptations in both hosts and parasites. For instance, there can be parasite manipulation of host behavior whereby parasites alter the behavior of their host in a way that increases their own transmission. In contrast, hosts may engage in self-medication, a series of behaviors through which animals exploit the anti-parasitic properties of a third species to prevent or reduce infection. Parasitic manipulation and self-medication both lead to substantial host behavioral changes with contrasting consequences on disease dynamics. Here, I will present some experimental results on parasitic manipulation in vector-borne diseases and on self-medication in monarch butterflies and will discuss their potential effects on disease transmissions.
Photo: Jaap de Roode
 Laura Pollittpolitt
Laura Pollittpolitt
Ph.D. student, Institute of Evolutionary Biology, University of Edinburgh
The evolutionary ecology of transmission strategies: from hosts to vectors
 Within-host infection dynamics have important implications for infectious disease biology, but the influence of the within-host environment on these dynamics is poorly understood. My research applies existing frameworks from social evolution, life history theory and phenotypic plasticity to understand malaria parasite traits and their interactions with co-infecting parasites and the within-host environment. I then integrate parasite within-host dynamics to the implications for transmission and successful progression through the vector.
Within-host infection dynamics have important implications for infectious disease biology, but the influence of the within-host environment on these dynamics is poorly understood. My research applies existing frameworks from social evolution, life history theory and phenotypic plasticity to understand malaria parasite traits and their interactions with co-infecting parasites and the within-host environment. I then integrate parasite within-host dynamics to the implications for transmission and successful progression through the vector.
Using experimental infection data, I will show that while in the vertebrate host, malaria parasites detect and respond to the presence of competitors by altering their transmission strategies to maximise survival. What’s more, these strategies are fine tuned to subtle differences in the within-host environment, including the availability of resources and a parasite’s relative competitive ability. These parasite strategies dramatically influence the number of transmission stages produced and taken up by an insect vector, where there are further important interactions with the environment. I show that high parasite densities in the mosquito vector lead to significant costs to parasite fitness (transmission) in terms of parasite development and vector survival. These results demonstrate that, across hosts and vectors, parasite strategies have significant implications for the transmission and severity of disease.
Photo: Sinclair Stammers
 Amber Smith
Amber Smith
Postdoctoral Research Associate, Theoretical Biology and Biophysics, Los Alamos National Laboratory
Bacterial coinfections with influenza: quantitative data and modeling
 Influenza associated secondary bacterial infections are a leading cause of death and have been identified as a major source of illness during the 1918 and 1957 influenza pandemics. Experimental studies suggest a lethal synergism between influenza and certain bacteria, particularly Streptococcus pneumoniae, but the precise processes involved are unclear. To address the mechanisms and determine the influences of pathogen dose and strain, we first infected groups of mice with influenza A virus then seven days later with S. pneumoniae. Surprisingly, following bacterial colonization, viral titers rebound initially before declining in infected mice. Bacterial titers rapidly rise to high levels that remain elevated. We use a mathematical model to explore the coupled interaction and expose the dominant phenomenological controlling mechanisms. We hypothesize that viral titers rebound with bacterial presence due to enhanced viral release from infected cells, and that bacterial titers increase due to alveolar macrophage impairment. Distinct dynamics are evident between initial bacterial dose but not with expression of the influenza 1918 PB1-F2 protein. Our proposed model provides a basis for the mathematical formulation of other processes that may address the interactions of viruses and bacteria during a dual infection.
Influenza associated secondary bacterial infections are a leading cause of death and have been identified as a major source of illness during the 1918 and 1957 influenza pandemics. Experimental studies suggest a lethal synergism between influenza and certain bacteria, particularly Streptococcus pneumoniae, but the precise processes involved are unclear. To address the mechanisms and determine the influences of pathogen dose and strain, we first infected groups of mice with influenza A virus then seven days later with S. pneumoniae. Surprisingly, following bacterial colonization, viral titers rebound initially before declining in infected mice. Bacterial titers rapidly rise to high levels that remain elevated. We use a mathematical model to explore the coupled interaction and expose the dominant phenomenological controlling mechanisms. We hypothesize that viral titers rebound with bacterial presence due to enhanced viral release from infected cells, and that bacterial titers increase due to alveolar macrophage impairment. Distinct dynamics are evident between initial bacterial dose but not with expression of the influenza 1918 PB1-F2 protein. Our proposed model provides a basis for the mathematical formulation of other processes that may address the interactions of viruses and bacteria during a dual infection.
Photo: Amber Smith
 Daniel Streickerstreicker
Daniel Streickerstreicker
Ph.D. Candidate, University of Georgia, Odum School of Ecology
Effects of host species identity on the epidemiology and evolutio n of bat rabies
n of bat rabies
Most pathogens studied to date are capable of infecting multiple host species. Because host species differ in their ecological, behavioral and physiological traits, it follows that pathogen transmission will depend on the particular host species infected. Using a dataset of hundreds of rabies viruses collected from 31 bat species, I explore the effects of host species identity on two key factors associated with viral emergence: cross-species transmission and rapid evolution. First, using a population genetic framework, I quantify the frequency of rabies transmission between bat species and reconstruct patterns of successful viral establishment across hosts. These estimates demonstrate that both cross-species transmission and host shifts decrease in frequency with increasing phylogenetic distance between bat species, indicating that the strength of host-species barriers may determine the fate of emerging host-virus interactions. Next, using phylogenetic comparative analysis, I consider how ecological differences among bats influence the speed of viral evolution. Results suggest that seasonal inactivity of bats forces an annual pause in viral replication and transmission that slows rates of rabies virus evolution, implying that rabies virus utilizes fundamentally different strategies for long term persistence in different hosts species. Together, these results provide unique insight into how host ecology and evolution can influence the dynamics and emergence of multi-host pathogens.
Photo: Daniel Streicker


