 RNase P catalyzes the maturation of the 5’ end of tRNA precursors. Typically these enzymes are ribonucleoproteins (RNPs) with a conserved RNA component responsible for catalysis. However, protein-only RNase P (PRORP) enzymes process precursor tRNAs in human mitochondria and all tRNA-encoded compartments of plants. Our work focuses on understanding how this novel class of RNase P enzymes functions by using techniques such as: steady-state and transient kinetics, mutagenesis, x-ray crystallography, crosslinking, and chemical probing of RNA. Our goal in the Fierke Lab is to provide insights into the catalytic strategies utilized by RNA and proteins that catalyze the same reaction, to gain insight into RNA maturation in mitochondria, and to understand how disease-related mutations in human mitochondrial tRNA affect RNase P processing.
RNase P catalyzes the maturation of the 5’ end of tRNA precursors. Typically these enzymes are ribonucleoproteins (RNPs) with a conserved RNA component responsible for catalysis. However, protein-only RNase P (PRORP) enzymes process precursor tRNAs in human mitochondria and all tRNA-encoded compartments of plants. Our work focuses on understanding how this novel class of RNase P enzymes functions by using techniques such as: steady-state and transient kinetics, mutagenesis, x-ray crystallography, crosslinking, and chemical probing of RNA. Our goal in the Fierke Lab is to provide insights into the catalytic strategies utilized by RNA and proteins that catalyze the same reaction, to gain insight into RNA maturation in mitochondria, and to understand how disease-related mutations in human mitochondrial tRNA affect RNase P processing.
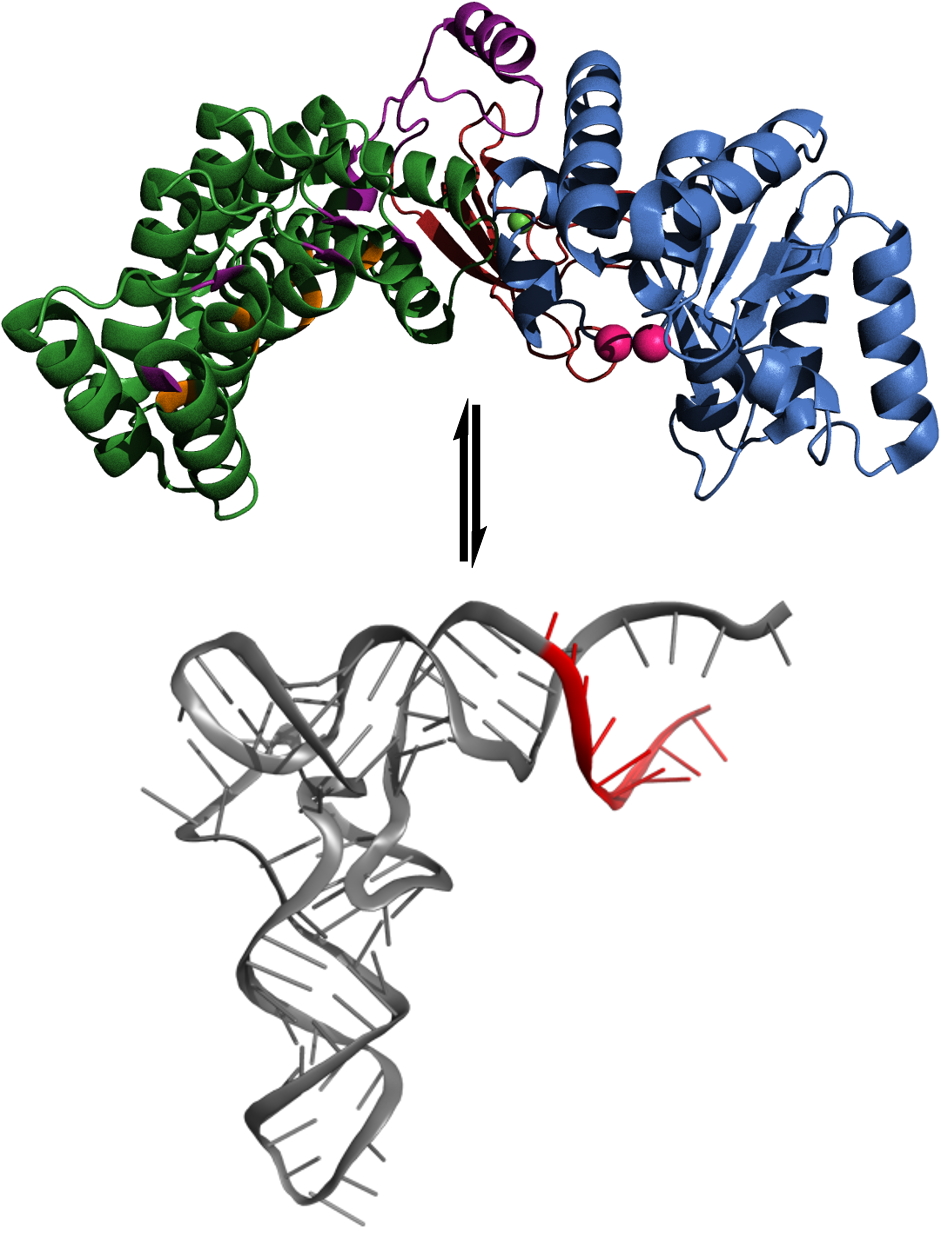
Using a high-throughput fluorescence polarization (FP) assay, we have identified both non-competitive and time-dependent inhibitors against A. thaliana mitochondrial RNase P (PRORP1) from a small compound library. These compounds will aid in therapeutic development for mitochondria-associated diseases. The PRORPs from plants, trypanosomes, and algae are single-subunit enzymes capable of efficient catalysis in vitro, but the PRORPs from humans require additional subunits for efficient catalysis. Both plant and human enzymes contain pentatricopeptide repeat (PPR) domains that are usually implicated in single-stranded RNA binding. The PRORP PPRs have non-canonical binding sites and likely utilize alternative means for recognition. Through mutagenesis and in vitro binding and catalytic activity assays we seek to understand the recognition of tRNA substrates. Recently, we have discovered an alternative PPR recognition strategy used by PRORPs to recognize their highly structured pre-tRNA substrates. Additionally, we have demonstrated that a methyltransferase complex (MRPP1-MRPP2) enhances substrate binding affinity for efficient catalysis by human RNase P metallonuclease subunit (MRPP3).
Collaborators on this project:
Markos Koutmos

