Networks are everywhere! Networks go beyond typically-used averages to provide an understanding of how brain regions “talk” to each other, how a person’s emotions are intertwined across a day, and how children influence one another during play. Thus, significant insight into neural, behavioral, and social systems can be garnered by determining how the parts of those systems interrelate. Network models require the collection and analysis of time series data, or data that are collected via intensive longitudinal measurement; examples include functional neuroimages, daily diaries, and coded observations. Group iterative multiple model estimation (GIMME), which implements unified structural equation models (uSEMs), is a valid and reliable network modeling technique and the primary connectivity method used in the lab; for a tutorial, see Beltz and Gates (2017). Extensions of this technique include the modeling of event external to the network, such as task effects in fMRI studies (Beltz, 2018), searches for multiple solutions (Beltz & Molenaar, 2016), and procedures for ensuring temporal information in the networks is accurately modeled (Beltz & Molenaar, 2015).
For slides and video of Dr. Beltz’s GIMME workshop visit the UM Methods Hour website.
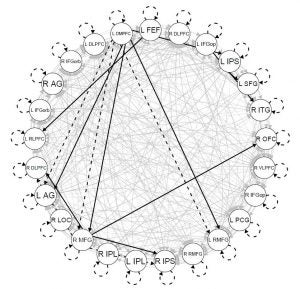
A map generated using GIMME showing connections among 25 brain regions of interest for a single participant during resting state. The connections are lagged (dashed lines), contemporaneous (solid lines), constant across development (black lines), or specific to key periods in development (gray lines).


