Overview
Our research is aimed at understanding the dynamic process that allows bacteria to integrate and adapt to their environment. The approach we’ve taken to this problem is to better understand the tools that microorganisms use to interact with themselves and their surroundings. This has led us to pursue extracellular organelle biology with special emphasis on curli fiber biogenesis by E. coli. During the course of our studies, we found that curli share distinguishing features with the medically important pathogenic amyloid fibers that are the hallmark of many neurodegenerative diseases such as Alzheimer’s, Huntington’s, systemic amyloidosis and the prion diseases.

Protein Misfolding Done Right
The discovery of a natural amyloid protein in E. coli opens up the possibility of performing genetic and biochemical experiments that are impractical or impossible in other amyloid model systems. Our work focuses on three related questions: how curli are assembled on the cell surface, how curli modify community behavior in bacteria, and how curli expression affects the host-pathogen interaction during an infection. We blend microscopy, biochemistry, and genetics in a concerted effort to delineate the structural, functional, and molecular details of curli, a distinct class of bacterial fibers that share properties with amyloid proteins. The curli biogenesis apparatus is possibly the world’s most tractable model system for understanding amyloid formation. But more than that, teasing apart the details of curli biogenesis will give insights into such fundamental bacterial processes as gene regulation, protein secretion and protein folding.
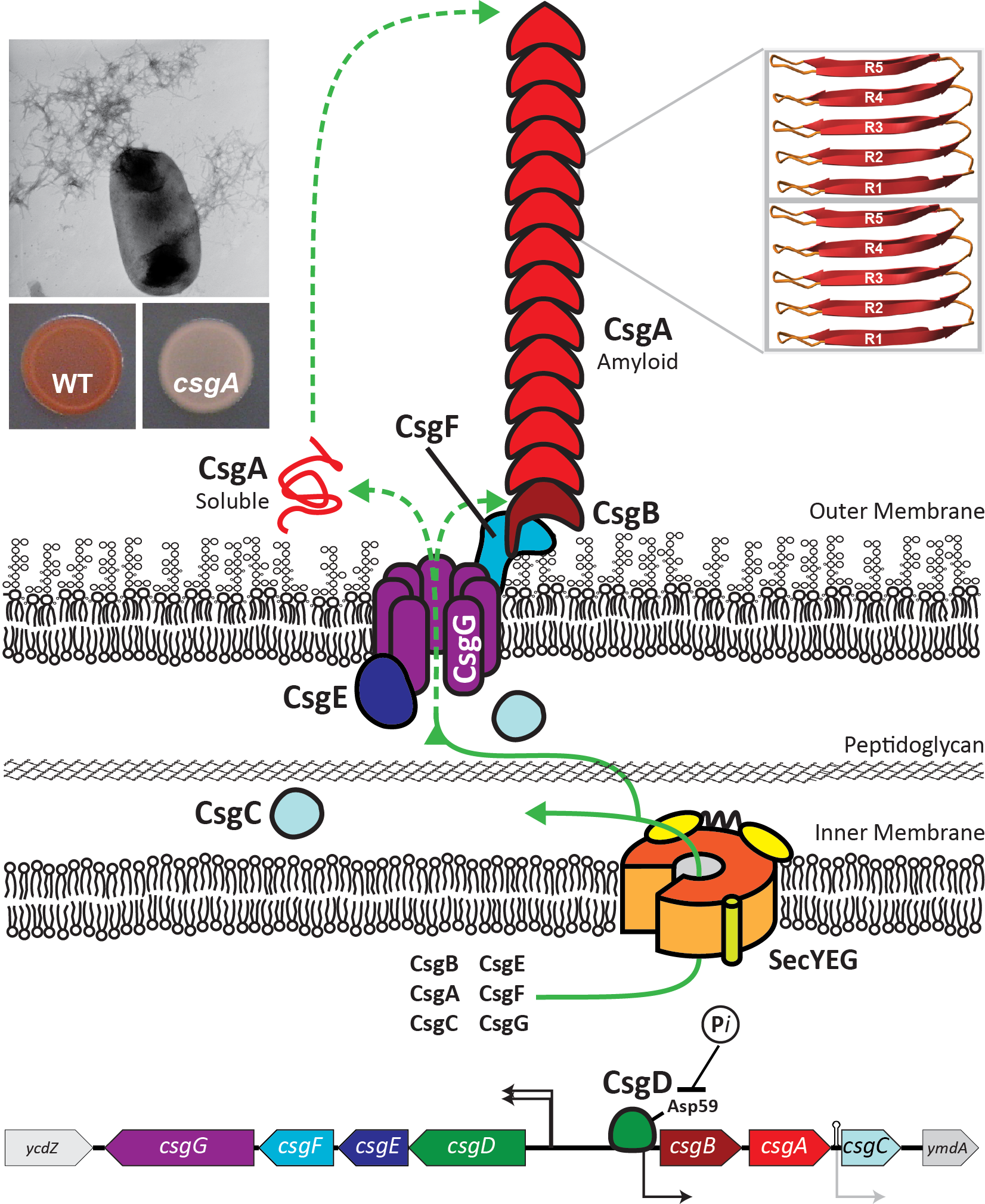
Curli Biogenesis
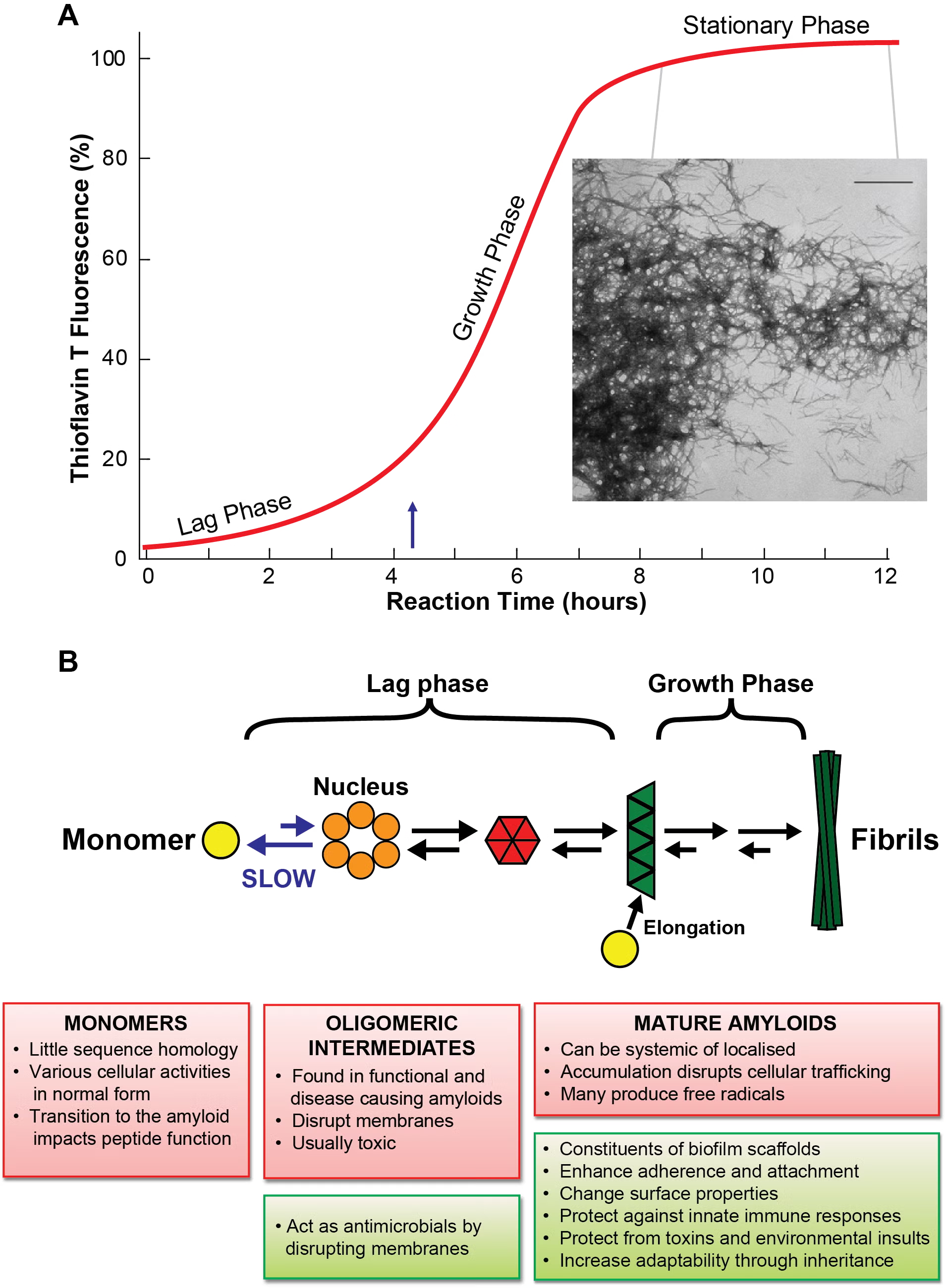
Much of the Chapman lab is interested in figuring out how curli amyloid fibers are assembled on the cell surface. Aggregation-prone curli subunits must be chaperoned from the inside of the cell to the outside, where they polymerize into stable amyloid fibers.
Protein and Chemical Chaperones
We are actively characterizing molecules that can ameliorate amyloid aggregation. E. coli has figured out how to coordinate amyloid formation so that happens at the correct time and the correct place. Helping to orchestrate bacterial amyloid formation are several chaperone-like proteins that discourage protein aggregation inside the cell. One of these proteins, called CsgC, works with remarkable efficiency, and without the need for ATP hydrolysis. We are trying to understand how the CsgC family of chaperones work and how we might apply the CsgC anti-amyloid activity to human proteins.
Amyloid formation can also be interrogated with chemical chaperones. Along with the Almqvist lab in Sweden, we are investigating the ability of rationally-designed 2-pyridone molecules to alter amyloid formation. These molecules are being tested as anti-amyloid/anti-biofilm reagents.
Biology of the Bacterial Extracellular Matrix
Cells in a biofilm are surrounded by an extracellular matrix that protects them from various stresses ranging from antibiotics to the host immune response. Resistance conferred by the extracellular matrix makes biofilm infections difficult to eradicate. Our lab is actively investigating the molecular pathways that control the extracellular matrix components, along with the role of the individual matrix components in biofilm resistance.
Protein Economics

We are also interested in the evolutionary pressures on proteins.
Evolution is easily appreciated to work in functional terms. Mutations that improve or diversify a protein’s function are selected for, whereas disruptive mutations are selected against. However, economy can also play a role in protein evolution. Read all about it in Dan Smith’s really cool paper: Economical Evolution: Microbes Reduce the Synthetic Cost of Extracellular Proteins


