We are using a combination of structural biology, biochemistry, cell biology and proteomics to investigate the in vivo and in vitro function of our new heat shock proteins.
Biochemistry
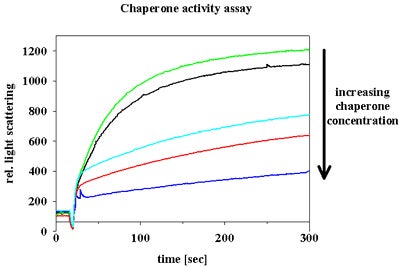
We work with purified protein components and we use spectroscopic techniques to analyze the activity of our proteins, to investigate metal binding properties, to monitor complex formation and to study conformational changes in our proteins. We analyze enzyme mechanisms and chaperone functions.
Cell Biology
We use the powerful tools of E. coli genetics to study the function of our proteins in vivo. We construct knock out strains and identify phenotypes under normal and stress conditions.
E. coli wild type cells can swarm to reach nutrients. In certain E. coli mutants, the flagellar motor is impaired and the E. coli cells lose their ability to swim.
Proteomics
We are using proteomics to study the effects of heat shock and oxidative stress on proteins and cells. 2D gel analysis is performed in our lab and mass spectroscopy is performed by the proteomics consortium of the University of Michigan.
Structures
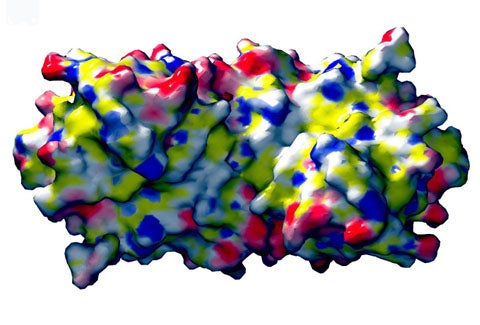
In collaboration with the crystallography group of Dr. Mark Saper at the University of Michigan, we have solved the high resolution crystal structure of Hsp33 and RrmJ.
Potential binding site for protein folding intermediates in Hsp33. The solvent contact surface with electrostatic potential is shown below.
Contact
Biological Sciences Building
Room 5022
1105 N. University
Ann Arbor, MI 48109
USA
ph | 734.615.1286
lab ph | 734.647.6683
fax | 734.647.0884
News
- U-M researchers discover stress in early life extends lifespan
- Versatile protein helps cells survive oxidative stress
- ICT News Desk: How Bleach Kills Bacteria
- Order out of Disorder: Working Cycle of an Intrinsically Unfolded Chaperone Cell
- The Secret Life of Bleach