Our thinking about gene regulation is dominated by biochemistry, yet the DNA containing our genes is a polymeric molecule whose mechanical properties need to be carefully considered. For certain regulatory processes such as DNA looping, that require contortions of the DNA, the mechanics might actively participate in controlling expression. We have been studying Lac repressor-mediated DNA looping as a model system for understanding biomechanical gene regulation.
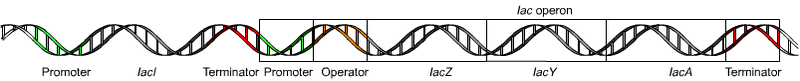
The lac operon is one of the better understood models of gene regulation and is responsible for the efficient metabolism of lactose within E. coli bacteria. To turn off expression when it is not needed, repressor protein binds to operator sites along the DNA. The protein can simultaneously bind to two operator sites forming a loop in the DNA, which prevents the polymerase from transcribing the gene.
DNA loops pose a tricky problem to a biologist primarily because they are out of reach of both NMR and crystallography. While there are crystal structures of the repressor protein in complex, the loop structures remain unresolved. Theoretical studies have shown that DNA loops can form in different topologies. Further, in an ideal in vitro situation, various topologies are found to coexist. Our lab tries to understand looping and the consequences of it through a variety of single-molecule techniques.